Volume 24, No.4 Pages 383 - 387
1. 最近の研究から/FROM LATEST RESEARCH
Long-term Proposal Report 2
Application & Development of Nuclear Resonance Vibrational Spectroscopy (NRVS) and Synchrotron Mossbauer Spectroscopy of Iron-Hydrogen Interactions in Hydrogenases, Nitrogenases, and Model Complexes
[1]SETI Institute、[2]University of California、[3]Diffraction and Scattering Division, Center for Synchrotron Radiation Research, JASRI、[4]Technische Universität Berlin、[5]Max Planck Institute for CEC、[6]Institute of Chemistry, Academia Sinica、[7]Department of Chemistry, University of Illinois
- Abstract
- Using 57Fe nuclear resonant vibrational spectroscopy (NRVS), we have characterized 57Fe-labeled proteins such as [FeFe] hydrogenase ([FeFe] H2ase), [NiFe] hydrogenase ([NiFe] H2ase), and nitrogenase (N2ase). Following the successful NRVS observation of the Ni-H-Fe mode in the Ni-R form of [NiFe] H2ase, we worked to extend this observation to the more difficult Ni-C species. We also continued studies of the [FeFe] H2ase from different organisms: Chlamydomonas reinhardtii (Cr-HydA1) and Desulfovibrio desulfuricans [FeFe] H2ase (Dd-HydAB). Fe-hydride and Fe-deuteride vibrational modes in [FeFe] H2ase were observed and interpreted by DFT calculations. We also observed the importance of the interaction of the amine ligand of the azadithiolate (ADT) bridge by comparing wild-type enzyme results with spectra for enzyme substituted with 13CD2-ADT. Model compounds have played an important role in interpreting these spectra, and papers were published on compounds with bridging hydrides or deuterides.
Background and Purpose
H2ases catalyze the reversible reaction of 2H+ + 2e- ⇄ H2[1-3][1] J. Y. H. Kim and H. J. Cha: Korean J. Chem. Eng. 30 (2013) 1-10.
[2] B. E. Jugder, J. Welch, K. F. Aguey-Zinsou and C. P. Marquis: Rsc. Advances 3 (2013) 8142-8159.
[3] W. Lubitz, H. Ogata, O. Rudiger and E. Reijerse: Chem. Rev. 114 (2014) 4081-4148., while N2ases catalyze the fixation of molecular nitrogen (N2) in the atmosphere into bio-available NH3[4,5][4] T. Spatzal, J. Schlesier, E.-M. Burger, D. Sippel, L. Zhang, S. L. A. Andrade, D. C. Rees and O. Einsle: Nature Comm. 7 (2016) 10902.
[5] B. M. Hoffman, D. Lukoyanov, D. R. Dean and L. C. Seefeldt: Acc. Chem. Res. 46 (2013) 587-595.. Since today's world faces multiple pressures from the demands for sustainable energy and food resources, H2ases and N2ases have both attracted a lot of attention and have been intensively studied for decades. Although crystal structures are available for all of these enzymes (Figure 1), many key enzyme intermediates cannot be crystallized. We are therefore using spectroscopy as an alternative probe of these key intermediates, with the overarching goal of understanding the catalytic mechanisms of these systems.
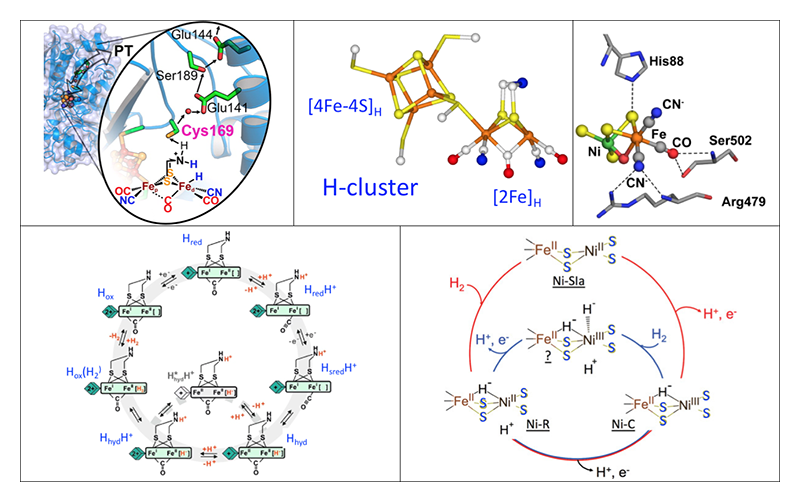
Figure 1 Top: Structures of the catalytic centers. Left to right: typical [FeFe] H2ase and the associated proton transfer chain; closeup of the active site H cluster; [NiFe] center inside a typical [NiFe] H2ase. Bottom: Proposed catalytic cycles. Left: [FeFe] H2ase, with key Hhyd intermediate. Right: [NiFe] H2ase, cycling between Ni-C, NiSIa, and Ni-R.
Nuclear resonant vibrational spectroscopy (NRVS) measures vibrational transitions that occur together with nuclear transitions that are typically associated with the Mossbauer effect[6-9][6] M. Seto, Y. Yoda, S. Kikuta, X. W. Zhang and M. Ando: Phys. Rev. Lett. 74 (1995) 3828-3831.
[7] W. Sturhahn, T. S. Toellner, E. E. Alp, X. Zhang, M. Ando, Y. Yoda, S. Kikuta, M. Seto, C. W. Kimball and B. Dabrowski: Phys. Rev. Lett. 74 (1995) 3832-3835.
[8] Y. Yoda, K. Okada, H. Wang, S. P. Cramer and M. Seto: Jap. J. App. Phys. 55 (2016) 122401.
[9] H. Wang, E. E. Alp, Y. Yoda and S. P. Cramer: Metalloproteins: Methods and Protocols 1122 (Springer, 2014) 125-138.. For the study of Fe in biology, 57Fe NRVS has key features that complement traditional techniques such as infrared (IR) and Raman spectroscopies. Despite the complexity of these samples, 57Fe NRVS only sees normal modes that involve motion of the 57Fe nucleus. Since the NRVS intensity is proportional to this 57Fe motion, the NRVS spectrum is easy to calculate from a normal mode analysis of a candidate structure. The technique has moderate sensitivity, and at the moment can be used to study frozen protein samples at ~mM concentrations[9,10][9] H. Wang, E. E. Alp, Y. Yoda and S. P. Cramer: Metalloproteins: Methods and Protocols 1122 (Springer, 2014) 125-138.
[10] M. C. Smith, Y. Xiao, H. Wang, S. J. George, D. Coucovanis, M. Koutmos, W. Sturhahn, E. E. Alp, J. Zhao and S. P. Cramer: Inorg. Chem. 44 (2005) 5562-5570.. Over the past decade, this technique has been used to study the lower frequency modes of a wide variety of Fe complexes and proteins[11-14][11] P. N. Serrano, H. Wang, J. C. Crack, C. Prior, M. I. Hutchings, A. J. Thomson, S. Kamali, Y. Yoda, J. Zhao, M. Y. Hu, E. E. Alp, V. S. Oganesyan, N. E. L. Brun and S. P. Cramer: Angew. Chem. Int. Ed. 55 (2016) 14575-14579.
[12] S. P. Cramer, Y. Xiao, H. Wang, Y. Guo and M. C. Smith: Hyperfine Interact. 170 (2006) 47-54.
[13] A. Scott, V. Pelmenschikov, Y. Guo, H. Wang, L. Yan, S. George, C. Dapper, W. Newton, Y. Yoda, Y. Tanaka and S. P. Cramer: J. Am. Chem. Soc. 136 (2014) 15942-15954.
[14] H. Ogata, T. Krämer, H. Wang, D. Schilter, V. Pelmenschikov, M. van Gastel, F. Neese, T. B. Rauchfuss, L. B. Gee, A. D. Scott, Y. Yoda, Y. Tanaka, W. Lubitz and S. P. Cramer: Nature Comm. 6 (2015) 7890.. More recently, we have extended NRVS studies to more difficult Fe-H related vibrational features in several H2ase enzymes as will be detailed later in this article[14][14] H. Ogata, T. Krämer, H. Wang, D. Schilter, V. Pelmenschikov, M. van Gastel, F. Neese, T. B. Rauchfuss, L. B. Gee, A. D. Scott, Y. Yoda, Y. Tanaka, W. Lubitz and S. P. Cramer: Nature Comm. 6 (2015) 7890..
Our NRVS measurements were performed at SPring-8 BL09XU[15][15] Y. Yoda, M. Yabashi, K. Izumi, X. W. Zhang, S. Kishimoto, S. Kitao, M. Seto, T. Mitsui, T. Harami, Y. Imai and S. Kikuta: Nucl. Inst. Meth. A 467-468 (2001) 715-718., which uses a high heat load monochromator to produce 14.4 keV radiation with ~1.0 eV resolution, followed by a Ge(422) × 2Si(975) high energy resolution monochromator (HRM) to narrow the resolution to ~0.8 meV, with a final flux of ~2.5 × 109 photons/s. Some NRVS data were also measured at SPring-8 BL19LXU. The NRVS measurements used a 2 × 2 avalanche photodiode (APD) detector array to collect the nuclear fluorescence and the internal conversion Fe Kα fluorescence following 57Fe nuclear excitation. The typical background count rates for this array is ~0.03 cts s-1. A closed-cycle liquid helium flow cryostat was used to maintain the samples at cryogenic temperatures (~50 K). During the NRVS measurements, the scans were often divided into segments with different data collection times at a given energy. These acquisition times ranged from 1-3 s in the Fe-S region to as much as 30 s per point for weak Fe-H modes. The energy scale was always calibrated with respect to a standard sample of [57FeCl4][NEt4], which has a prominent peak at 380 cm-1. NRVS data were converted to an 57Fe partial vibrational density of state (PVDOS) using the PHOENIX software package[7][7] W. Sturhahn, T. S. Toellner, E. E. Alp, X. Zhang, M. Ando, Y. Yoda, S. Kikuta, M. Seto, C. W. Kimball and B. Dabrowski: Phys. Rev. Lett. 74 (1995) 3832-3835..
Results
[FeFe] H2ases
Our collaborators in Mulheim have an established protocol for reconstitution of the apo-enzyme with synthetic mimics for the binuclear cluster. Using [57Fe2(odt)(CN)2(CO)4]2- prepared in the Rauchfuss lab (odt = oxadithiolate), they successfully replaced the amine at the bridge of the active site of Cr-HydA1 with an oxygen to form the ODT variant (Figure 2). This approach allowed us for the specific 57Fe labeling of the [2Fe]H subsites, while leaving other irons unlabeled. We then prepared a transient intermediate (known as the Hhyd state)[16][16] H. Long, P. W. King and C. H. Chang: J. Phys. Chem. B 118 (2014) 890-900., and we observed Fe-H bending modes at ~670 and ~730 cm-1 (Figure 2). The Fe-H assignment was observed by D exchange, which moved one band to ~625 cm-1 while the other feature became buried in the Fe-CO/CN region. This work was published in J. Am. Chem. Soc.[17][17] E. J. Reijerse, C. C. Pham, V. Pelmenschikov, R. Gilbert-Wilson, A. Adamska-Venkatesh, J. F. Siebel, L. B. Gee, Y. Yoda, K. Tamasaku, W. Lubitz, T. B. Rauchfuss and S. P. Cramer: J. Am. Chem. Soc. 139 (2017) 4306-4309..
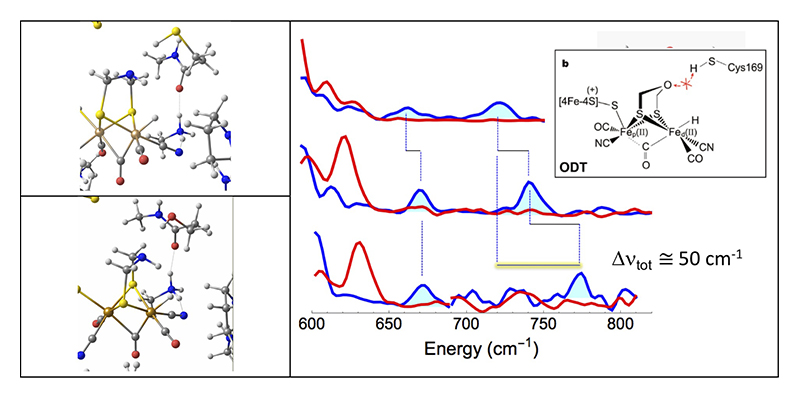
Figure 2 Left: DFT predictions for the active site Hhyd local structure for (top) the wild-type enzyme and (bottom) the C169S variant. Right: 57Fe PVDOS for various types CrHydA1 [FeFe] H2ase. Top to bottom: wild-type enzyme with ADT replaced by ODT; wild-type enzyme; C169S variant. Spectra are for H2/H2O (—) and D2/D2O (—) conditions. Inset: structure and inhibition mechanism of the ODT variant.
We were also able to observe the Hhyd intermediate in 57Fe-labeled wild type Cr-HydA1 and Dd-HydAB with the natural ADT at the active site. We inspected the Hhyd forms of both of these enzymes and found the Fe-H bands now shifted to ~675 and ~745 cm-1 (Figure 2). DFT calculations reproduced these shifts as a consequence of the presence of a proton on the ADT nitrogen. This work was also published in J. Am. Chem. Soc.[18][18] V. Pelmenschikov, J. A. Birrell, C. C. Pham, N. Mishra, H. X. Wang, C. Sommer, E. Reijerse, C. P. Richers, K. Tamasaku, Y. Yoda, T. B. Rauchfuss, W. Lubitz and S. P. Cramer: J. Am. Chem. Soc. 139 (2017) 16894-16902..
Finally, we turned to the C169S mutant of Cr-HydA1 where the Hhyd species was originally observed (Figure 2). The Hhyd species for this variant yielded bands at ~673 and ~772 cm-1 (Figure 2). The latter banded is shifted by ~45 - 50 cm-1 from the original observation on the ODT variant. DFT calculations were used to explain this shift in terms of variations in bond strengths and distanced along the serine-ADT-H-Fe network (Figure 2). This was published in Angew. Chem. Int. Ed.[19][19] C. C. Pham, D. W. Mulder, V. Pelmenschikov, P. W. King, M. W. Ratzloff, H. Wang, N. Mishra, E. E. Alp, J. Zhao, M. Y. Hu, K. Tamasaku, Y. Yoda and S. P. Cramer: Angew. Chem. Int. Ed. 57 (2018) 10605-10609..
[NiFe] H2ases
The local crystal structure for the [NiFe] center is shown in Figure 1. This enzyme has two species that have bridging hydrides between the Ni and Fe (Figure 3 - left). Previously, we were successful observing the Ni-H-Fe wag mode, as described article in Nature Comm. in 2015[14][14] H. Ogata, T. Krämer, H. Wang, D. Schilter, V. Pelmenschikov, M. van Gastel, F. Neese, T. B. Rauchfuss, L. B. Gee, A. D. Scott, Y. Yoda, Y. Tanaka, W. Lubitz and S. P. Cramer: Nature Comm. 6 (2015) 7890., (Figure 3 - right). Since then, we have been engaged in trying to see the same mode for Ni-C H2ase. This has turned out to be much more difficult. DFT calculations have shown that there is less Fe motion for the wag mode in this species. Work continues.
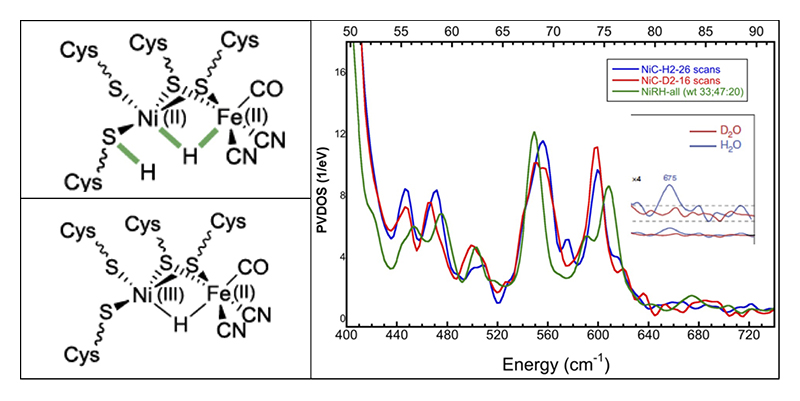
Figure 3 Left: proposed structures for Ni-R and Ni-C forms of [NiFe] H2ase. Right: NRVS for Ni-R and Ni-C forms. Inset: expanded Ni-H-Fe region for Ni-R H2ase.
Model Compounds
Interpretation of enzyme NRVS has been aided by model compound spectra. Structures of complexes and their NRVS are shown in Figure 4. All of this work has been published or submitted[20-23][20] M. R. Carlson, D. L. Gray, C. P. Richers, W. Wang, P.-H. Zhao, T. B. Rauchfuss, V. Pelmenschikov, C. C. Pham, L. B. Gee, H. Wang and S. P. Cramer: Inorg. Chem. 57 (2018) 1988-2001.
[21] V. Pelmenschikov, L. B. Gee, H. Wang, K. C. MacLeod, S. F. McWilliams, K. L. Skubi, S. P. Cramer and P. L. Holland: Angew. Chem. Int. Ed. 57 (2018) 9367-9371.
[22] F. Wittkamp, N. Mishra, H. Wang, H.-C. Wille, S. P. Cramer, U.-P. Apfel and V. Pelmenschikov: Chem. Sci. 10 (2019) 7535-7541.
[23] L. B. Gee, V. Pelmenschikov, H. Wang, N. Mishra, Y.-C. Liu, Y. Yoda, K. Tamasaku, M. Kaupp, M.-H. Chiang and S. P. Cramer: Chem. Int. Ed. (2019) submitted..
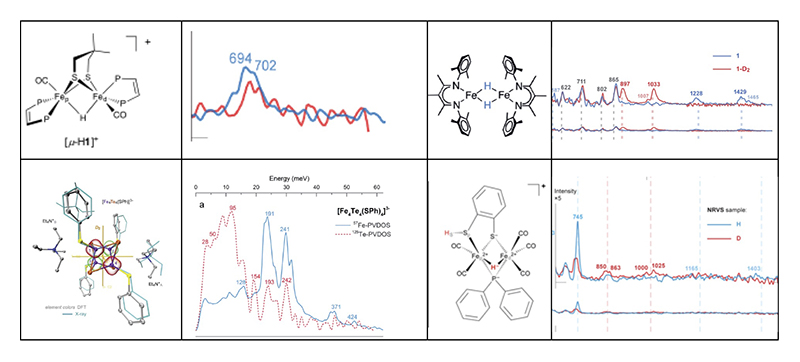
Figure 4 Top left pair: the bridging hydride isomer of a Rauchfuss model complex and the observed Fe-H-Fe mode in the NRVS[20][20] M. R. Carlson, D. L. Gray, C. P. Richers, W. Wang, P.-H. Zhao, T. B. Rauchfuss, V. Pelmenschikov, C. C. Pham, L. B. Gee, H. Wang and S. P. Cramer: Inorg. Chem. 57 (2018) 1988-2001.. Top right pair: a complex with two bridging hydrides from the Holland lab, and associated complex NRVS[21][21] V. Pelmenschikov, L. B. Gee, H. Wang, K. C. MacLeod, S. F. McWilliams, K. L. Skubi, S. P. Cramer and P. L. Holland: Angew. Chem. Int. Ed. 57 (2018) 9367-9371.. Lower left: an [Fe4Te4] cluster from Apfel lab and associated NRVS from Fe and Te points of view (Te data from PETRA-III)[22][22] F. Wittkamp, N. Mishra, H. Wang, H.-C. Wille, S. P. Cramer, U.-P. Apfel and V. Pelmenschikov: Chem. Sci. 10 (2019) 7535-7541.. Lower right: a bridging hydride from Chang lab and associated NRVS[23][23] L. B. Gee, V. Pelmenschikov, H. Wang, N. Mishra, Y.-C. Liu, Y. Yoda, K. Tamasaku, M. Kaupp, M.-H. Chiang and S. P. Cramer: Chem. Int. Ed. (2019) submitted..
Summary
Over this long-term proposal (2017A0141-2018B0141) at SPring-8 BL09XU, and in combination with some work from BL19LXU, we have obtained fruitful NRVS results for many important enzymes. In this article, we illustrated how NRVS can probe Fe-H/D modes that are hard to see by other techniques. SPring-8 remains the best synchrotron light source in the world for these experiments.
Acknowledgments
We thank Dr. Kenji Tamasaku very much for his assistance in our beamtime at BL19LXU and other contributions.
References
[1] J. Y. H. Kim and H. J. Cha: Korean J. Chem. Eng. 30 (2013) 1-10.
[2] B. E. Jugder, J. Welch, K. F. Aguey-Zinsou and C. P. Marquis: Rsc. Advances 3 (2013) 8142-8159.
[3] W. Lubitz, H. Ogata, O. Rudiger and E. Reijerse: Chem. Rev. 114 (2014) 4081-4148.
[4] T. Spatzal, J. Schlesier, E.-M. Burger, D. Sippel, L. Zhang, S. L. A. Andrade, D. C. Rees and O. Einsle: Nature Comm. 7 (2016) 10902.
[5] B. M. Hoffman, D. Lukoyanov, D. R. Dean and L. C. Seefeldt: Acc. Chem. Res. 46 (2013) 587-595.
[6] M. Seto, Y. Yoda, S. Kikuta, X. W. Zhang and M. Ando: Phys. Rev. Lett. 74 (1995) 3828-3831.
[7] W. Sturhahn, T. S. Toellner, E. E. Alp, X. Zhang, M. Ando, Y. Yoda, S. Kikuta, M. Seto, C. W. Kimball and B. Dabrowski: Phys. Rev. Lett. 74 (1995) 3832-3835.
[8] Y. Yoda, K. Okada, H. Wang, S. P. Cramer and M. Seto: Jap. J. App. Phys. 55 (2016) 122401.
[9] H. Wang, E. E. Alp, Y. Yoda and S. P. Cramer: Metalloproteins: Methods and Protocols 1122 (Springer, 2014) 125-138.
[10] M. C. Smith, Y. Xiao, H. Wang, S. J. George, D. Coucovanis, M. Koutmos, W. Sturhahn, E. E. Alp, J. Zhao and S. P. Cramer: Inorg. Chem. 44 (2005) 5562-5570.
[11] P. N. Serrano, H. Wang, J. C. Crack, C. Prior, M. I. Hutchings, A. J. Thomson, S. Kamali, Y. Yoda, J. Zhao, M. Y. Hu, E. E. Alp, V. S. Oganesyan, N. E. L. Brun and S. P. Cramer: Angew. Chem. Int. Ed. 55 (2016) 14575-14579.
[12] S. P. Cramer, Y. Xiao, H. Wang, Y. Guo and M. C. Smith: Hyperfine Interact. 170 (2006) 47-54.
[13] A. Scott, V. Pelmenschikov, Y. Guo, H. Wang, L. Yan, S. George, C. Dapper, W. Newton, Y. Yoda, Y. Tanaka and S. P. Cramer: J. Am. Chem. Soc. 136 (2014) 15942-15954.
[14] H. Ogata, T. Krämer, H. Wang, D. Schilter, V. Pelmenschikov, M. van Gastel, F. Neese, T. B. Rauchfuss, L. B. Gee, A. D. Scott, Y. Yoda, Y. Tanaka, W. Lubitz and S. P. Cramer: Nature Comm. 6 (2015) 7890.
[15] Y. Yoda, M. Yabashi, K. Izumi, X. W. Zhang, S. Kishimoto, S. Kitao, M. Seto, T. Mitsui, T. Harami, Y. Imai and S. Kikuta: Nucl. Inst. Meth. A 467-468 (2001) 715-718.
[16] H. Long, P. W. King and C. H. Chang: J. Phys. Chem. B 118 (2014) 890-900.
[17] E. J. Reijerse, C. C. Pham, V. Pelmenschikov, R. Gilbert-Wilson, A. Adamska-Venkatesh, J. F. Siebel, L. B. Gee, Y. Yoda, K. Tamasaku, W. Lubitz, T. B. Rauchfuss and S. P. Cramer: J. Am. Chem. Soc. 139 (2017) 4306-4309.
[18] V. Pelmenschikov, J. A. Birrell, C. C. Pham, N. Mishra, H. X. Wang, C. Sommer, E. Reijerse, C. P. Richers, K. Tamasaku, Y. Yoda, T. B. Rauchfuss, W. Lubitz and S. P. Cramer: J. Am. Chem. Soc. 139 (2017) 16894-16902.
[19] C. C. Pham, D. W. Mulder, V. Pelmenschikov, P. W. King, M. W. Ratzloff, H. Wang, N. Mishra, E. E. Alp, J. Zhao, M. Y. Hu, K. Tamasaku, Y. Yoda and S. P. Cramer: Angew. Chem. Int. Ed. 57 (2018) 10605-10609.
[20] M. R. Carlson, D. L. Gray, C. P. Richers, W. Wang, P.-H. Zhao, T. B. Rauchfuss, V. Pelmenschikov, C. C. Pham, L. B. Gee, H. Wang and S. P. Cramer: Inorg. Chem. 57 (2018) 1988-2001.
[21] V. Pelmenschikov, L. B. Gee, H. Wang, K. C. MacLeod, S. F. McWilliams, K. L. Skubi, S. P. Cramer and P. L. Holland: Angew. Chem. Int. Ed. 57 (2018) 9367-9371.
[22] F. Wittkamp, N. Mishra, H. Wang, H.-C. Wille, S. P. Cramer, U.-P. Apfel and V. Pelmenschikov: Chem. Sci. 10 (2019) 7535-7541.
[23] L. B. Gee, V. Pelmenschikov, H. Wang, N. Mishra, Y.-C. Liu, Y. Yoda, K. Tamasaku, M. Kaupp, M.-H. Chiang and S. P. Cramer: Chem. Int. Ed. (2019) submitted.








