Volume 20, No.3 Pages 241 - 245
1. 最近の研究から/FROM LATEST RESEARCH
Long-term Proposal Report
Structural Study of a Bacterial Homologue of SWEET Transporters
Center for Structural Biology, School of Medicine and School of Life Sciences, Tsinghua University
- Abstract
Sugar transporters mediate sugar transport across biological membranes and have crucial roles in plants, worms and animals. They are essential for the maintenance of animal blood glucose level, plant nectar production and pollen development. Genes encoding sugar transporters have been identified, functionally expressed and studied in the past decades. At present, three classes of eukaryotic sugar transporters have been characterized, including the glucose transporters (GLUTs), sodium-glucose symporters (SGLTs), and bidirectional sugar transporter SWEETs. In humans, understanding of sugar transport has therapeutic importance. For example, GLUTs in human being can transport glucose across the plasma membrane, maintain blood glucose level and supply energy for cell survival and growth. In plants, these transporters are important for crop yield and pathogen invasion. SWEETs represent a novel sugar transport family that was first identified in plants. They contain seven transmembrane segments (TMs) and selectively transport monosaccharides such as glucose and fructose or disaccharides such as sucrose across plasma or intracellular membranes. Homologues of SWEETs were recently identified in bacteria. Each bacterial SWEET monomer consists of three TMs, similar to one three-helix bundle in the eukaryotic SWEETs. Therefore, they were named SemiSWEETs. Here we report the crystal structure of a SemiSWEET.
Introduction
Sugar is the major source of energy and is an important carbon skeletons composition of the organism. Sugar transporting plays important roles for cell growth and survival in plants, worms and animals. They are essential for the maintenance of animal blood glucose level, plant nectar production, seed and pollen development. A lot of key components both prokaryotic and eukaryotic encoding sugar transporters have been identified, functionally expressed and studied in the past years. At present, three classes of eukaryotic sugar transporters have been characterized. They are the glucose transporters (GLUTs), sodium-glucose symporters (SGLTs), and SWEETs. GLUTs belongs to the major facilitator superfamily (MFS), and is related with many diseases, like cancer. SGLTs transport substrates using a different way compared with GLUTs, which co-transport sodium during transporting. SWEETs belongs to a novel family of membrane sugar transporters that have been identified in plants, worms, and mammals. They can selectively transport mono- or disaccharides over plasma or intracellular membranes, and are also involved in a number of important physiological processes[1][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694.. The functions of SWEETs are best characterized in plants. Members of this family show diverse substrate selectivity. In Arabidopsis thaliana, AtSWEET1/4/5/7/8/13 mediate glucose efflux[1][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694., AtSWEET11/12 function as sucrose transporters[1][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694., and AtSWEET17 permeates fructose[2][2] L. Q. Chen et al., "Sugar transporters for intercellular exchange and nutrition of pathogens" Nature 2010 468 (7323) 527-532.. These SWEETs are important for the growth and development of plants, and some are hijacked by pathogens or symbionts for their own sugar supply.
To study the sugar transport mechanism, we need to get high resolution structures both with and without different substrates. Atomic resolution structure also could help us find the pathogenic changes and provide the therapeutic potentials of intractable diseases like diabetes. For this reason, a lot of work had done on this project. Dr. Yan’s group solved the fucose transporter FucP in Escherichia coli in 2010[3][3] S. Dang et al., "Structure of a fucose transporter in an outward-open conformation" Nature 2010 467 (7316) 734-738. and a bacterial homologue of glucose transporters GLUT1-4 XylE in 2012[4][4] L. Sun et al., "Crystal structure of a bacterial homologue of glucose transporters GLUT1-4" Nature 2012 490 (7420) 361-366.. More importantly, Dr. Yan’s group solved the human GLUT1 structure in 2014[5][5] D. Deng et al., "Crystal structure of the human glucose transporter GLUT1" Nature 2014 510 (7503) 121-125.. They all belongs to MFS superfamily. These structures provided clues to understand substrate binding and transport mechanism on MFS superfamily, however, there are many other questions remain unknown:
1. What are the transport mechanism of other sugar transporters like SWEETs superfamily?
2. What are the structure and functional mechanism of the substrate binding and specificity?
To answer these questions, we proposed to:
1. To understand the transport mechanism of SWEETs;
2. To determine the structure of SWEETs with/without different substrates, and test the substrate specificity using biochemical and biophysical method.
It’s not easy to solve all the problems especially for membrane proteins. Therefore, for a long-term proposal with SPring-8, we expect to make progresses on the structure determination of SWEETs, and on the same time try to go further on the substrate binding and specificity regulation.
Progress
SWEETs belong to the MtN3 family in plants and SLC50 sugar efflux transporter family in human[1][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694.. It was predicted that there are seven transmembrane (TM) helices in SWEETs which are folded into two parallel three-helix bundles connected by one central TM[1,2][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694.
[2] L. Q. Chen et al., "Sugar transporters for intercellular exchange and nutrition of pathogens" Nature 2010 468 (7323) 527-532.. Recently, the bacterial homologues of SWEETs were identified by bioinformatic analysis[1][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694.. Each bacterial SWEET monomer consists of three TMs, similar as the three-helix bundles in the eukaryotic SWEETs. Therefore they are called SemiSWEETs. The representative homologue from B. japonicum USDA 110, BjSemiSWEET1, showed sucrose transport activity, just like AtSWEET11[1][1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694..
In an attempt to understand the molecular basis underlying substrate selectivity and transport mechanism of sugar transporters, we sought to determine the crystal structure of SemiSWEETs. We determined the crystal structure of the SemiSWEET from T. yellowstonii DSM 11347 (TySemiSWEET), a close homologue of BjSemiSWEET in an occluded state at 2.5 Å resolution in the space group P212121 using lipidic cubic phase approach with data screened and collected at BL41XU and BL32XU, SPring-8. The structure of TySemiSWEET was determined by molecular replacement using the recently reported structure of a SemiSWEET protein from L. biflexa (LbSemiSWEET)[6][6] Y. Xu et al., "Structures of bacterial homologues of SWEET transporters in two distinct conformations" Nature 2014 515 (7527) 448-452. as search model and refined to 2.4 Å resolution.
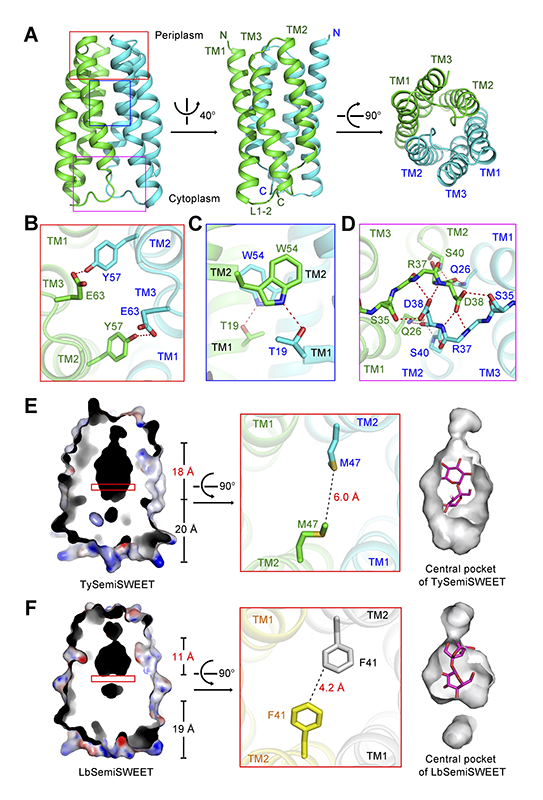
In each asymmetric unit, two TySemiSWEET molecules assemble into a dimer (Figure 1A). The two TySemiSWEETs are almost identical to each other. In each TySemiSWEET molecule, the transmembrane helix 1 (TM1) and TM2 are linked by the linker L1-2 containing seven amino acid residues. TM2 shows about 24° rotation relative to TM1 (Figure 1A). TM3 folds between TM1 and TM2. According to the 'inside-positive rule' and the topology of AtSWEET11, it was predicted that the linker L1-2 locates at the cytosolic side of TySemiSWEET (Figure 1A)[6,7][6] Y. Xu et al., "Structures of bacterial homologues of SWEET transporters in two distinct conformations" Nature 2014 515 (7527) 448-452.
[7] N. Yan, "Structural advances for the major facilitator superfamily (MFS) transporters" Trends Biochem Sci 2013 38 (3) 151-159..
Figure 1 Crystal structure of the SemiSWEET from T. yellowstonii (TySemiSWEET) in an occluded conformation. (A) Overall structure of the dimeric TySemiSWEET. The two protomers are colored green and cyan. (B-D) The dimer interface of TySemiSWEET consists of three clusters of H-bonds between residues on TM1 of one protomer and TM2 of the other, including a pair of H-bonds at the extracellular side (B), a pair close to the center of the membrane (C), and an extensive H-bond network on the cytoplasmic side (D). The H-bonds, together with extensive van der Waals contacts between the two protomers, sealed the dimer in an occluded conformation. (E-F) The central pocket of TySemiSWEET is considerably larger than that of LbSemiSWEET. Residues Phe41 from the two protomers of LbSemiSWEET close the central pocket at approximately the midway of the membrane, whereas the corresponding Met47 residues in TySemiSWEET leave enough space for an elongated pocket. A sucrose molecule can be accommodated by TySemiSWEET, but not LbSemiSWEET (right panels). All structure figures were prepared with PyMol[10][10] W. L. DeLano, The PyMOL Molecular Graphics System on World Wide Web (http://www.pymol.org) 2002..
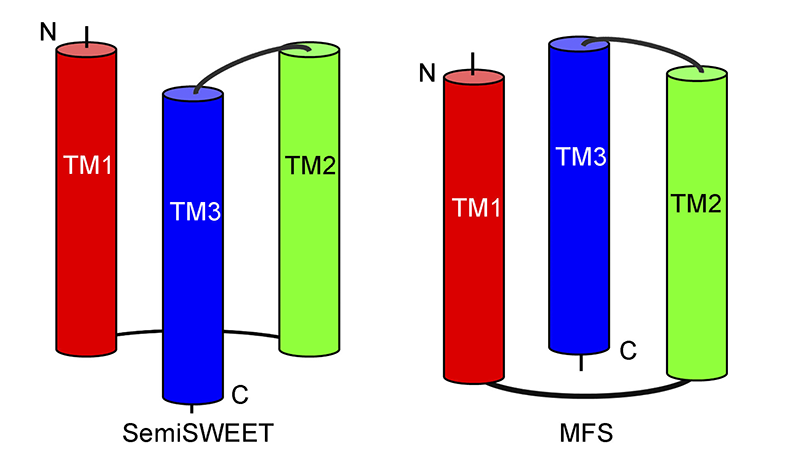
As we know that the basic structural and functional unit of MFS transporters is also the 3-helix bundle[7][7] N. Yan, "Structural advances for the major facilitator superfamily (MFS) transporters" Trends Biochem Sci 2013 38 (3) 151-159.. It’s interesting that in the TySemiSWEET and MFS structures, aligned TM1 and TM2 in each 3-helix bundle on the same plane, TM3 in SemiSWEETs is on the opposite side to that of the MFS (Figure 2). This may suggest that there’s a different ancestor between SWEET and MFS transporters.
Figure 2 Topology of the 3-helix bundle in SemiSWEET and MFS. SemiSWEET shows a similar but not identical topology in compared with the 3-helix bundle in MFS. In SemiSWEET, the transmembrane (TM) helix TM3 locates in front of the surface defined by TM1 and TM2. In contrast, TM3 locates behind the surface defined by TM1 and TM2 in the 3-helix bundle in MFS.
The two protomers in each dimer enclose a central pocket that is sealed from both sides of the membrane. Therefore, the structure represents an occluded conformation. The dimer interface is mediated through three clusters of hydrogen bonds (H-bonds) between TM1 of one protomer and TM2 and linker 2-3 of the other (Figure 1B, 1C and 1D) as well as van der Waals contacts.
The sequence identities of TySemiSWEET with BjSemiSWEET and LbSemiSWEET is 44.2% and 40.2%, respectively. The structure of LbSemiSWEET was also determined in an occluded conformation[6][6] Y. Xu et al., "Structures of bacterial homologues of SWEET transporters in two distinct conformations" Nature 2014 515 (7527) 448-452.. However, evident difference can be observed in the central pockets of the two highly similar structures (Figure 1E and 1F). The central pocket of TySemiSWEET is 18 Å long, with an overall surface of 463 Å2 and volume of 613 Å3. In contrast, that of LbSemiSWEET is 11 Å long, with an overall surface of 327 Å2 and volume of 424 Å3. The difference is caused mainly by variation of one amino acid, Met47 in TySemiSWEET versus the corresponding Phe41 in LbSemiSWEET. The bulky side chains of the two Phe41 in LbSemiSWEET dimer close the central pocket in the midway of the membrane, whereas Met47 residues in TySemiSWEET leave enough space for an elongated central pocket (Figures 1E, 1F).
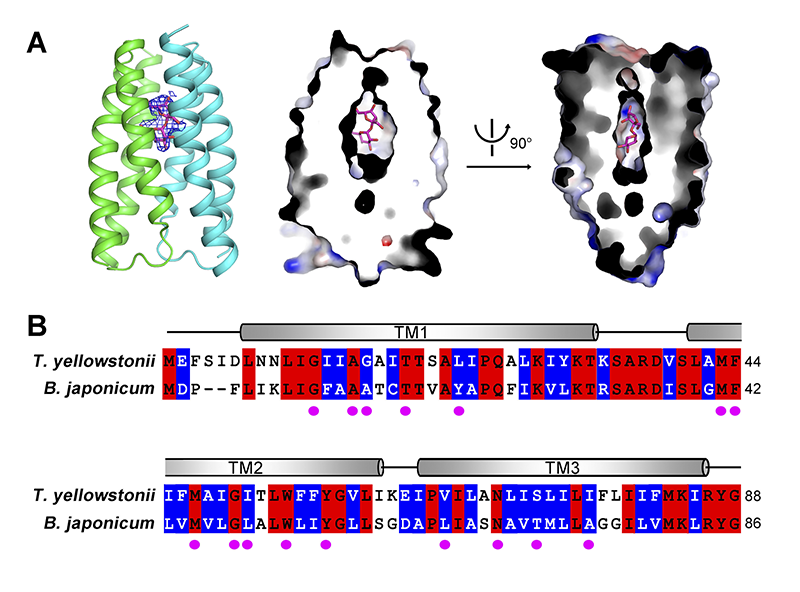
Furthermore, there is an electron density in the central pocket after structural refinement of TySemiSWEET. Although the density is not in atomic resolution, we can’t identify it accurately, the crystals of TySemiSWEET were obtained in the presence of 20 mM sucrose, and the contour of the electron density is similar to a disaccharide molecule (Figure 3A). Therefore, we tentatively built a sucrose molecule into it. The disaccharide molecule can be fit well by the surrounding residues in the central pocket (Figures 1E). What’s more, all the 16 residues forming the central pocket are highly conserved between TySemiSWEET and BjSemiSWEET, which means there’s a similar central pocket with BjSemiSWEET, the sucrose transporter (Figure 3B). In contrast, the pocket in LbSemiSWEET is too small to accommodate a disaccharide molecule, consistent with its function of being a glucose transporter (Figure 1F, right panel).
Figure 3 Sucrose is the potential substrate of TySemiSWEET. (A) Docking of one molecule of sucrose in the central pocket of TySemiSWEET. There is an obvious electron density at the center of the TySemiSWEET dimer. The identity of this electron density cannot be determined at this resolution. However, one molecule of sucrose can be built in, and matches the pocket well. (B) Sequence alignment between TySemiSWEET and BjSemiSWEET. The residues identical in the two proteins are colored red; the residues with a similar hydrophobicity are colored blue. The magenta spots indicate the residues that are placed towards the central pocket in TySemiSWEET.
Future plan
Structural comparison of TySemiSWEET and LbSemiSWEET provides important clues to understand substrate binding and selectivity of SemiSWEETs. The structures of the outward-open and occluded states have obtained for the SemiSWEETs[6][6] Y. Xu et al., "Structures of bacterial homologues of SWEET transporters in two distinct conformations" Nature 2014 515 (7527) 448-452.. Later, another paper reported SemiSWEET structure from Escherichia coli, in the both inward-open and outward-open states[8][8] Y. Lee et al., "Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter" Nat Commun 2015 6 6112.. But till now none of the available SemiSWEETs structures are bound with any definite substrates, although all the structures seems to have the same putative substrate-binding pocket[6,8,9][6] Y. Xu et al., "Structures of bacterial homologues of SWEET transporters in two distinct conformations" Nature 2014 515 (7527) 448-452.
[8] Y. Lee et al., "Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter" Nat Commun 2015 6 6112.
[9] J. Wang et al., "Crystal structure of a bacterial homologue of SWEET transporters" Cell Res 2014 24 (12) 1486-1489.. Therefore more structural and biochemical work are needed. Other than that, there remains other questions to be characterized like: What’s the driving force for their conformational changes? Whether SemiSWEETs and SWEETs are facilitative uniporters or secondary active co-transporters? Is the eukaryotic SWEETs function in the same way? In spite of this, the structures we reported here and previously lay out the foundation to address some of important questions. And we believe that our later work will unveil more in the near future.
Annotation
The figures and some of the progress description were adopted from the published paper by Wang et al.[9][9] J. Wang et al., "Crystal structure of a bacterial homologue of SWEET transporters" Cell Res 2014 24 (12) 1486-1489..
The results were published as a letter on Cell Res[9][9] J. Wang et al., "Crystal structure of a bacterial homologue of SWEET transporters" Cell Res 2014 24 (12) 1486-1489..
References
[1] Y. H. Xuan et al., "Functional role of oligomerization for bacterial and plant SWEET sugar transporter family" Proc Natl Acad Sci USA 2013 110 (39) E3685-3694.
[2] L. Q. Chen et al., "Sugar transporters for intercellular exchange and nutrition of pathogens" Nature 2010 468 (7323) 527-532.
[3] S. Dang et al., "Structure of a fucose transporter in an outward-open conformation" Nature 2010 467 (7316) 734-738.
[4] L. Sun et al., "Crystal structure of a bacterial homologue of glucose transporters GLUT1-4" Nature 2012 490 (7420) 361-366.
[5] D. Deng et al., "Crystal structure of the human glucose transporter GLUT1" Nature 2014 510 (7503) 121-125.
[6] Y. Xu et al., "Structures of bacterial homologues of SWEET transporters in two distinct conformations" Nature 2014 515 (7527) 448-452.
[7] N. Yan, "Structural advances for the major facilitator superfamily (MFS) transporters" Trends Biochem Sci 2013 38 (3) 151-159.
[8] Y. Lee et al., "Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter" Nat Commun 2015 6 6112.
[9] J. Wang et al., "Crystal structure of a bacterial homologue of SWEET transporters" Cell Res 2014 24 (12) 1486-1489.
[10] W. L. DeLano, The PyMOL Molecular Graphics System on World Wide Web (http://www.pymol.org) 2002.