Volume 19, No.3 Pages 225 - 229
1. 最近の研究から/FROM LATEST RESEARCH
長期利用課題報告
Three-dimensional Analysis of Neuronal Circuits of the Human Brain by X-ray Microtomography
[1]東海大学 工学部/大学院工学研究科/大学院総合理工学研究科 Department of Applied Biochemistry, Tokai University (Corresponding author) 、[2]東海大学 工学部 School of Engineering, Tokai University、[3](公財)高輝度光科学研究センター 利用研究促進部門 Research & Utilization Division, JASRI
- Abstract
- Neuronal circuits are responsible for brain functions including verbal ability, reasoning, emotion, and decision-making. Neurons constitute neuronal circuits by building a three-dimensional network in brain tissue. The functional mechanisms of the human brain can therefore be revealed by unveiling the three-dimensional structure of human brain tissue. Here, we report on three-dimensional analysis of neuronal circuits of the human brain by synchrotron-radiation X-ray microtomography.
1. Introduction
You, the readers of this report, might exert your verbal ability to understand the meaning of this sentence, analyze the intent of the context, take an interest in it or a dislike to it, or be looking for something useful for your own research. These are all functions of the human brain[1][1] 水谷隆太、雑賀里乃:放射光 26 (2013) 268-275..
It has not been revealed how these elaborative functions, such as verbal ability, reasoning, emotion, and decision-making, are implemented in the human brain. Yet the tide of studies concerning the functional mechanisms of the brain is rising rapidly. For example, the Ministry of Education, Culture, Sports, Science and Technology of Japan granted the Strategic Research Program for Brain Sciences (started in 2008) 19.0 billion JPY for seven years and recently launched another project for the investigation of neuronal circuit functions of throughout primate brains by using innovative technologies (6.1 billion JPY for FY2013-2014). The National Institute of Health of the United States conducted the Human Connectome Project from 2009 to 2014 (38.5 million USD for five years); this project was recently succeeded by the BRAIN Initiative (100 million USD for FY2014). The European Union established the Human Brain Project in 2013 (to receive 1.2 billion Euros in the coming ten years). The inspiration behind this research trend is the successful experience of the Human Genome Project, which led to a revolution in the biological sciences. It has been suggested that interdisciplinary efforts should be made to respond to such expectations[2][2] T. R. Insel, S. C. Landis and F. S. Collins: Science 340 (2013) 687-688.. But it seems that these projects will take years to analyze the neuronal circuits of the human brain at the cellular level.
In order to determine the brain circuits composed of individual neurons, it will be essential to visualize the three-dimensional structure of brain tissue at subcellular resolution and also to identify each constituent in the three-dimensional map of the tissue structure. With this in mind, we have performed structural studies of human brain tissue with synchrotron-radiation X-ray microtomography (micro-CT)[3-6][3] R. Mizutani, A. Takeuchi, K. Uesugi, M. Ohyama, S. Takekoshi et al.: Brain Res. 1199 (2008) 53-61.
[4] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Tissue Eng. C14 (2008) 359-363.
[5] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Cerebral Cortex 20 (2010) 1739-1748.
[6] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, N. Nakamura et al.: AIP Conf. Proc. 1365 (2011) 403-406.. A skeletonized model of the human brain network of frontal cortex tissue was built by tracing the three-dimensional distribution of X-ray linear attenuation coefficients[6][6] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, N. Nakamura et al.: AIP Conf. Proc. 1365 (2011) 403-406.. Neuronal circuits were analytically resolved from the skeletonized model[5][5] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Cerebral Cortex 20 (2010) 1739-1748.. Although many studies have been reported on the visualization of neural tissues, ours is the only report that describes human brain circuits composed of individual neurons, as far as we know.
2. Visualization of brain tissue structure
A huge number of neurons constitute neuronal circuits by forming a network in brain tissue. Because the brain resides in three-dimensional space, its circuits can be revealed by analyzing this three-dimensional structure. We performed circuit analyses by (1) preparing tissue samples, (2) acquiring microtomographic datasets, (3) tracing the network structure in the resultant three-dimensional maps, and (4) determining the neuronal circuits from the skeletonized model (Figure 1).
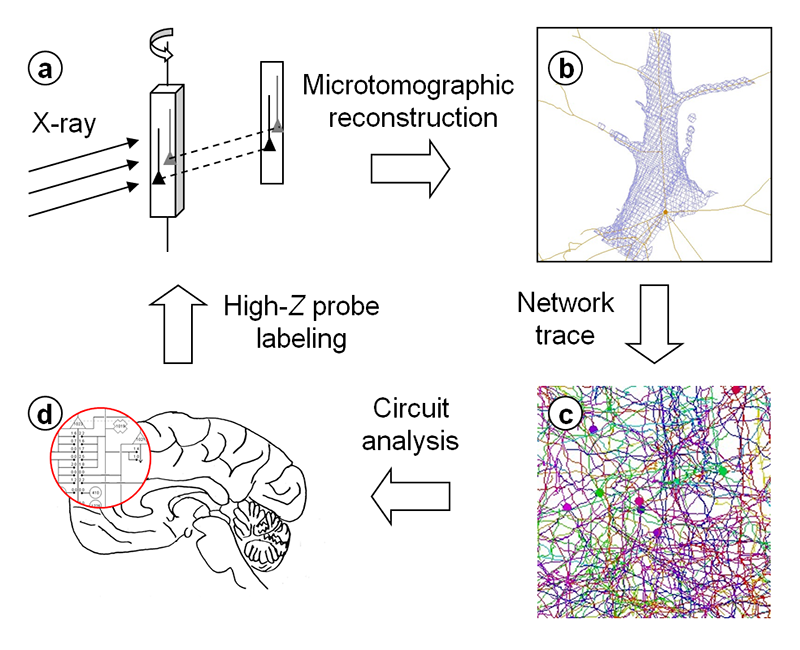
Figure 1. Schematic representation of X-ray microtomographic analysis of human brain circuits. Three-dimensional structure of human brain tissue labeled with high-Z elements was determined by microtomography (a). The obtained three-dimensional maps were used for network tracing (b), giving the structural model (c). Neuronal circuits were determined from the model (d). Modified from R. Mizutani: SPring-8 Research Frontiers (JASRI, 2010) p. 42.
Biological soft tissues, including those of the human brain, are composed of light elements that show little interaction with hard X-rays. Even if slight differences in physical density were to be visualized with contrast-sensitive methods (such as phase contrast techniques), the obtained structure would not necessarily be biologically relevant. In light microscopy or electron microscopy, target structures are labeled with dyes that interact with visible light or an electron beam. Therefore, in X-ray visualization, the structure of interest should be highlighted by labeling target constituents with high atomic-number (high-Z) elements that effectively interact with X-rays. Several methods for labeling biological tissues with high-Z elements have been reported[7][7] R. Mizutani and Y. Suzuki: Micron 43 (2012) 104-115.. We have recently reported X-ray visualization of biological samples by introducing a metalloprotein gene[8][8] R. Mizutani, K. Taguchi, M. Ohtsuka, M. Kimura, A. Takeuchi et al.: J. Synchrotron Radiat. 20 (2013) 581-586.. In this study, we applied the Golgi method to human brain tissue in order to visualize neuronal structures. The labeled tissues were embedded in epoxy resin in order to prevent structural fluctuations of the brain tissue during the acquisition of the X-ray images. This resin embedding process was essential for analyzing the three-dimensional structure at micrometer to submicrometer resolution. The obtained samples were then subjected to synchrotron-radiation X-ray microtomography at the BL20XU and BL47XU beamlines of the SPring-8 facility. Tomographic sections were reconstructed from the acquired images with the convolution back-projection method implemented in the RecView program (available from http://www.el.u-tokai.ac.jp/ryuta/). The spatial resolution of the obtained three-dimensional image was estimated by using test objects prepared by focused ion-beam milling[9-11][9] R. Mizutani, A. Takeuchi, K. Uesugi and Y. Suzuki: J. Synchrotron Radiat. 15 (2008) 648-654.
[10] R. Mizutani, K. Taguchi, A. Takeuchi, K. Uesugi and Y. Suzuki: Nucl. Inst. Meth. A621 (2010) 615-619.
[11] R. Mizutani, A. Takeuchi, R. Y. Osamura, S. Takekoshi, K. Uesugi et al.: Micron 41 (2010) 90-95.. Figure 2 shows an example of the three-dimensional structure of human brain tissue.

Figure 2. Three-dimensional structure of human cerebral tissue. The brain surface is toward the top. The skeletonized model shown in Figure 4 was built from this structure.
3. Analysis of human brain circuits
The human brain network was visualized as a three-dimensional image, i.e., a three-dimensional distribution of observed values. Its structure is too complicated to comprehend the entire constituents at a glance, as seen in Figure 2. Therefore, the image had to be further analyzed to reveal the neuronal circuits embedded in it.
Similar examples of three-dimensional analysis of complicated structures can be found in the field of X-ray crystallography, by which three-dimensional structures of biological macromolecules including proteins and DNAs have been elucidated. In crystallography, electron density maps are calculated from diffraction amplitudes, and then molecular models described in three-dimensional coordinate space are built by tracing electron density maps (Figure 3). The resultant models are used for discussing chemical mechanisms or molecular interactions. Although the target of crystallography is completely different from that of our study, the three-dimensional structure of brain tissue can be analyzed with a similar procedure (Figure 3). In our study, neuronal network models were built by placing and connecting nodes in the three-dimensional map. Neuronal circuits can be resolved from this skeletonized model.
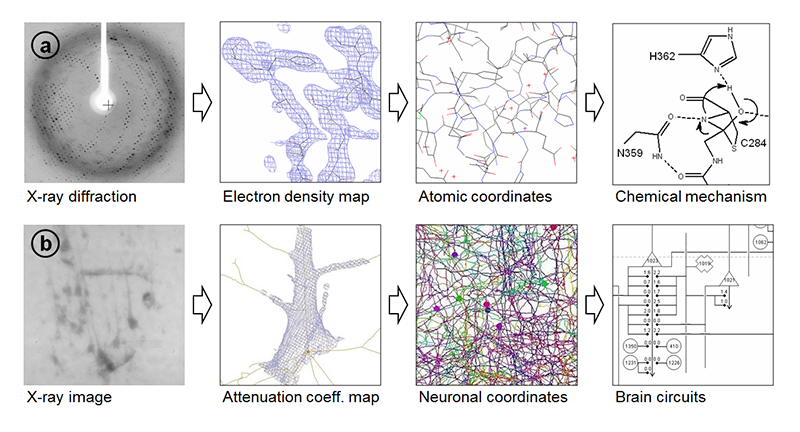
Figure 3. (a) In crystallography, observed diffractions are subjected to a Fourier transform to generate electron density maps. Atomic coordinates are then determined by analyzing the map. The resultant model is used for discussing chemical mechanisms or molecular interactions[17][17] R. Mizutani, S. Nogami, M. Kawasaki, Y. Ohya, Y. Anraku et al.: J. Mol. Biol. 316 (2002) 919-929.. (b) In microtomography, the observed images are subjected to tomographic reconstruction, yielding a three-dimensional map. Neuronal network structures are built by placing and connecting nodes in the map. The resultant model can be used to determine brain circuits[5][5] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Cerebral Cortex 20 (2010) 1739-1748.. Modified from R. Mizutani et al.: Microscopy: Science, Technology, Applications and Education, (Formatex Research Centre, 2010) p. 379.
However, it takes considerable man-hours to build three-dimensional models of neuronal networks. Therefore, automatic tracing should be incorporated into the model building[6,12][6] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, N. Nakamura et al.: AIP Conf. Proc. 1365 (2011) 403-406.
[12] Y. Liu: Neuroinformatics 9 (2011) 99-102.. We found that the Sobel operator[13][13] K. A. Al-Kofahi, S. Lasek, D. H. Szarowski, C. J. Pace, G. Nagy et al.: IEEE Trans. Inf. Technol. Biomed. 6 (2002) 171-187. and gradient vector flow methods[14][14] C. Xu and J. L. Prince: IEEE Trans. Image Process. 7 (1998) 359-369. are appropriate for tracing the neuronal networks of the human brain. These methods were implemented in dedicated software to obtain the initial models of the neuronal networks[6][6] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, N. Nakamura et al.: AIP Conf. Proc. 1365 (2011) 403-406.. The initial models were then manually examined and edited by using graphical interfaces. An example of the skeletonized model of the human brain network is shown in Figure 4. This model was built from the structure shown in Figure 2.
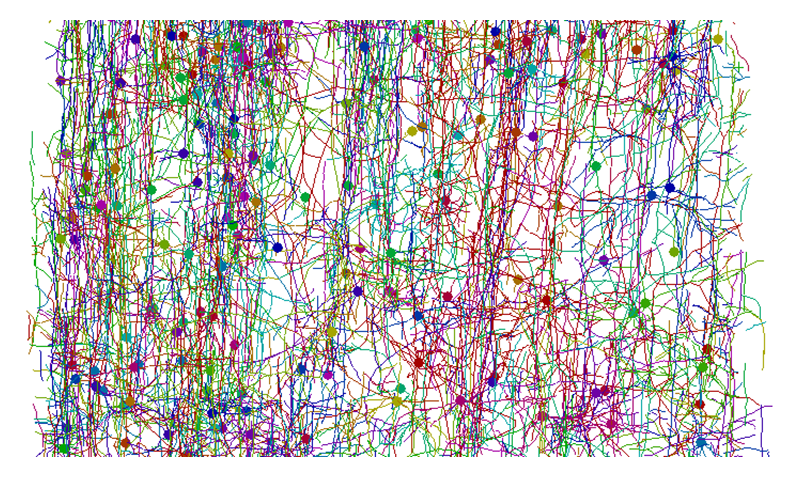
Figure 4. Skeletonized model of neuronal network of human cerebral cortex shown in Figure 2. Skeletonized models of neurons built by tracing three-dimensional distribution of linear attenuation coefficients are color-coded. Closed circles indicate soma locations. The brain surface is toward the top.
The obtained models are represented with three-dimensional Cartesian coordinates. Each structural element is assigned to individual neurons, capillary vessels, and so on. Although the network structure looks complicated, the distances between structural constituents can be readily calculated from the three-dimensional coordinates. This allows us to determine the neuronal circuits in the brain tissue by analyzing the positional relationships of the neurons[5][5] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Cerebral Cortex 20 (2010) 1739-1748.. Some of the circuits form feedback loops, in which inputs and outputs are interconnected with each other within a certain group of neurons. These circuits look like the flip-flops or multivibrators of electronics. Such structures seem to be common in the analyzed structures and are likely fundamental elements of human brain circuits.
4. Prospects of brain circuit analysis
We still have only a limited number of examples of human-brain neuronal circuits from a limited number of cases. Dependence on brain region, differences between human individuals, and changes due to psychiatric diseases remain to be clarified. We have already revealed the three-dimensional network of an entire hemisphere of a Drosophila brain[15][15] R. Mizutani, R. Saiga, A. Takeuchi, K. Uesugi and Y. Suzuki: J. Struct. Biol. 184 (2013) 271-279.. In some far-distant future, the entire circuit of the human brain will be able to be subjected to structural analysis. Such a study would lead to the simulation and reconstruction of human brain functions. The challenge after the determination of the entire circuit is reverse engineering of the machine language executed in the human brain. The functional mechanisms of the brain should be able to be revealed by taking these steps.
Analysis of brain circuits is an emerging science in the post-genomic era. Studies regarding human brain circuits will lay a scientific foundation for better diagnostics and therapeutics for those who suffer from mental disorders[2][2] T. R. Insel, S. C. Landis and F. S. Collins: Science 340 (2013) 687-688.. The Ministry of Health, Labour and Welfare of Japan announced that, from 2013, mental disorders are to be included in five major diseases that should be covered in the Medical Care Plan[16][16] Annual Health, Labour, and Welfare Report 2011-2012. Ministry of Health, Labour and Welfare.. However, drugs for psychiatric diseases are being administrated without a precise understanding of their mechanisms of action on neuronal circuits. This state of affairs is due to the limitations of our knowledge about human brain circuits. We have to squarely address the issue of analysis of human brain circuits. Such an analysis should provide a scientific underpinning for satisfying unmet medical needs of psychiatric patients.
Acknowledgements and Notes
This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (nos. 21611009, 25282250, and 25610126). The synchrotron radiation experiments were performed at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal nos. 2011A0034-2013B0034, and 2011B0041-2013B0041). Some parts of this article were translated from reference[1][1] 水谷隆太、雑賀里乃:放射光 26 (2013) 268-275. under the permission of the Japan Society for Synchrotron Radiation Research. Studies using human tissues were performed according to the terms admitted by the ethical committees of related organizations.
References
[1] 水谷隆太、雑賀里乃:放射光 26 (2013) 268-275.
[2] T. R. Insel, S. C. Landis and F. S. Collins: Science 340 (2013) 687-688.
[3] R. Mizutani, A. Takeuchi, K. Uesugi, M. Ohyama, S. Takekoshi et al.: Brain Res. 1199 (2008) 53-61.
[4] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Tissue Eng. C14 (2008) 359-363.
[5] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, R. Y. Osamura et al.: Cerebral Cortex 20 (2010) 1739-1748.
[6] R. Mizutani, A. Takeuchi, K. Uesugi, S. Takekoshi, N. Nakamura et al.: AIP Conf. Proc. 1365 (2011) 403-406.
[7] R. Mizutani and Y. Suzuki: Micron 43 (2012) 104-115.
[8] R. Mizutani, K. Taguchi, M. Ohtsuka, M. Kimura, A. Takeuchi et al.: J. Synchrotron Radiat. 20 (2013) 581-586.
[9] R. Mizutani, A. Takeuchi, K. Uesugi and Y. Suzuki: J. Synchrotron Radiat. 15 (2008) 648-654.
[10] R. Mizutani, K. Taguchi, A. Takeuchi, K. Uesugi and Y. Suzuki: Nucl. Inst. Meth. A621 (2010) 615-619.
[11] R. Mizutani, A. Takeuchi, R. Y. Osamura, S. Takekoshi, K. Uesugi et al.: Micron 41 (2010) 90-95.
[12] Y. Liu: Neuroinformatics 9 (2011) 99-102.
[13] K. A. Al-Kofahi, S. Lasek, D. H. Szarowski, C. J. Pace, G. Nagy et al.: IEEE Trans. Inf. Technol. Biomed. 6 (2002) 171-187.
[14] C. Xu and J. L. Prince: IEEE Trans. Image Process. 7 (1998) 359-369.
[15] R. Mizutani, R. Saiga, A. Takeuchi, K. Uesugi and Y. Suzuki: J. Struct. Biol. 184 (2013) 271-279.
[16] Annual Health, Labour, and Welfare Report 2011-2012. Ministry of Health, Labour and Welfare.
[17] R. Mizutani, S. Nogami, M. Kawasaki, Y. Ohya, Y. Anraku et al.: J. Mol. Biol. 316 (2002) 919-929.
東海大学工学部/大学院工学研究科/大学院総合理工学研究科
〒259-1292 神奈川県平塚市北金目4-1-1
TEL : 0463-58-1211
e-mail : ryuta@tokai-u.jp
東海大学工学部
〒259-1292 神奈川県平塚市北金目4-1-1
TEL : 0463-58-1211
(公財)高輝度光科学研究センター 利用研究促進部門
〒679-5198 兵庫県佐用郡佐用町光都1-1-1
TEL : 0791-58-0833
e-mail : take@spring8.or.jp
(公財)高輝度光科学研究センター 利用研究促進部門
〒679-5198 兵庫県佐用郡佐用町光都1-1-1
TEL : 0791-58-0833
e-mail : ueken@spring8.or.jp
(公財)高輝度光科学研究センター 利用研究促進部門
〒679-5198 兵庫県佐用郡佐用町光都1-1-1
TEL : 0791-58-0833
e-mail : yoshio@spring8.or.jp








