Volume 19, No.1 Pages 12 - 17
1. 最近の研究から/FROM LATEST RESEARCH
Long-term Proposal Report 2: Nuclear Resonance Vibrational Spectroscopy (NRVS) of Iron-Based Enzymes for Hydrogen Metabolism, Nitrogen Fixation, Small Molecule Sensing, DNA Repair, Photosynthesis, and Iron Storage
[1]Department of Chemistry, University of California - Davis, [2]Physical Biosciences Division, Lawrence Berkeley Laboratory, [3]Department of Biochemistry, Virginia Polytechnic Institute, [4]Max-Planck-Institut für Chemische Energiekonversion, [5]Department of Chemical Engineering, Stanford University, [6]School of Chemistry, University of East Anglia, [7]Research & Utilization Division, JASRI
Introduction
Nuclear resonant vibrational spectrascopy (NRVS) involves scanning monochromatic (~1 meV) x-rays through a nuclear resonance (in this case 57Fe) and monitoring transitions that correspond to vibrational modes. Over the latest phase of our program, we have used NRVS to monitor the reactions of small molecules with Fe-S proteins. The systems that we have examined include [FeFe] and [NiFe] hydrogenases that catalyze the conversions between H2 to electrons and protons, and nitrogenase, which converts N2 to NH3. We also work on Fe-S proteins that bacteria use as sensors of NO and O2.
O2 & NO 'Sensor' Progress
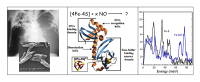
Fe-S clusters are important targets for NO-related physiological signal transduction[1][1] "Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator", Ding, H. G.; Demple, B. Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 5146-5150. as well as toxicity via interference with respiration and DNA replication[2][2] "Interplay between NO and Fe-S clusters: Relevance to biological systems", Drapier, J. C. Methods, 1997, 11, 319-329.. For example, the tuberculosis-causing bacterium Mycobacterium tuberculosis uses an Fe-S cluster protein to sense the presence of NO and O2 via its [4Fe-4S] cluster (Figure 1 left and middle). We have continued NRVS (Figure 1 right), EXAFS, and Mössbauer studies of NO reactivity with the WhiD proteins from the LeBrun lab. Our data showed that multiple NO ligands were bound to each Fe, but we found no evidence for intermediate species with substoichiometric amounts of NO. It appears that once the reaction with NO starts, subsequent decomposition steps are fast.
Figure 1. Left: evidence for tuberculosis in the lungs, and the bacteria that cause it. Middle: a typical Fe-S cluster sensor protein. Right: NRVS spectra for the natural (black) and NO treated WhiD proteins.
[FeFe] H2ase Progress
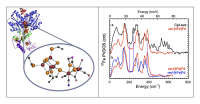
In the [FeFe] subset of H2ases, catalysis takes place at the so-called 'H-cluster', consisting of a [FeFe]H subcluster active site linked to a [4Fe-4S] cluster via a cysteine thiolate sulfur (Figure 2). The unique Fe subcluster site has multiple CO and CN– ligands, as well as a special bridging dithiolate with most likely N in the central position. In Clostridium pasteurianum H2ase I (CpI)[3][3] "X-ray Crystal Structure of the Fe-Only Hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom Resolution", Peters, J. W.; Lanzilotta, W. N.; Lemon, B. J.; Seefeldt, L. C. Science, 1998, 282, 1853-1858., Fe is also present in the 'proximal' [4Fe-4S] cluster that is part of the H-cluster, in an additional three [4Fe-4S] 'F-clusters', as well as a [2Fe-2S] cluster.
As seen in Figure 2, despite the presence of many other Fe-S clusters, by selective enrichment we were able to enhance the signal from the [2Fe] subcluster site. The characteristic bands in the 500-600 cm-1 range come from the multiple Fe-CO modes of this cluster. This work has been published in Biochemistry[4][4] "Nuclear Resonance Vibrational Spectroscopy and Electron Paramagnetic Resonance Spectroscopy of 57Fe-Enriched [FeFe] Hydrogenase Indicate Stepwise Assembly of the H-Cluster", Kuchenreuther, J. M.; Guo, Y.; Wang, H.; Myers, W. K.; George, S. J.; Boyke, C. A.; Yoda, Y.; Alp, E. E.; Zhao, J.; Britt, R. D.; Swartz, J. R.; Cramer, S. P. Biochemistry, 2013, 52, 818–826..
Figure 2. Left: The structure of the active site in [FeFe] H2ase. Right: (a) 57Fe PVDOS for 57Fe-enriched CpI [FeFe] H2ase (-) vs. oxidized D14C Pf (-); (b) 57Fe PVDOS for oxidized (-) vs. reduced (-) D14C Pf Fd.
[NiFe] H2ase Progress
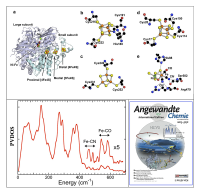
In the [NiFe] subset of H2ases, catalysis takes place at a Ni-Fe bimetallic center[5][5] "NiFe hydrogenases: structural and spectroscopic studies of the reaction mechanism", Ogata, H.; Lubitz, W.; Higuchi, Y. Dalton Trans., 2009, 37, 7577-7587.. In this active site, Fe is linked to Ni by a pair of cysteine thiolate ligands. The unique Fe site also has one CO and a pair of CN– ligands, making it the first organoiron center to be discovered in Nature[6][6] "Biological Activation of Hydrogen", Happe, R. P.; Roseboom, W.; Pierik, A. J.; Albracht, S. P. J.; Bagley, K. A. Nature, 1997, 385, 126.. [NiFe] H2ases invariably possess additional Fe-S clusters that relay electrons to and from the active site. In the case of Desulfovibrio vulgaris Miyazaki F., this chain consists of a conventional 'proximal' [4Fe-4S] cluster, a 'medial' [3Fe-4S] cluster, and a 'distal' [4Fe-4S] cluster with 3 cysteine ligands and a histidine ligand, as illustrated in Figure 3 (upper panel).
As illustrated in Figure 3 (lower left), when the 400-650 cm-1 region is magnified, peaks emerge that have intensities and frequencies consistent with Fe-CN and Fe-CO stretching and bending modes. In particular, the oxidized protein has two bands with maxima around 542 and 582 cm-1, that shift to ~ 547 and ~ 605 cm-1 in the reduced sample (reduced data not shown). These bands can be assigned to Fe-CO stretch and Fe-C-O bend modes, respectively. As frequently noted[7][7] "Quantitative Vibrational Dynamics of Iron in Carbonyl Porphyrins", Leu, B. M.; Silvernail, N. J.; Zgierski, M. Z.; Wyllie, G. R. A.; Ellison, M. K.; Scheidt, W. R.; Zhao, J.; Sturhahn, W.; Alp, E. E.; Sage, J. T. Biophys. J., 2007, 92, 3764-3783., strong Fe-CO backbonding pushes the Fe-CO bend mode (in this case at 582 cm-1) to a higher frequency than the Fe-CO stretch mode (in this case at 542 cm-1). Density functional theory (DFT) calculations have also shown that the bend mode mixes with a 'tilt' mode[8][8] "Carbonyl Tilting and Bending Potential Energy Surface of Carbon Monoxyhemes", Ghosh, A.; Bocian, D. F. J. Phys. Chem., 1996, 100, 6363-6367.. The small shifts to higher frequency with reduction are consistent with the well-known inverse correlation between νFeC and νCO in FeCO complexes[9-12][9] "Is Bound CO Linear or Bent in Heme-Proteins - Evidence from Resonance Raman and Infrared Spectroscopic Data", Li, X. Y.; Spiro, T. G. J. Am. Chem. Soc., 1988, 110, 6024-6033.
[10] "Connection between the taxonomic substates and protonation of histidines 64 and 97 in carbonmonoxy myoglobin", Müller, J. D.; McMahon, B. H.; Chien, E. Y. T.; Sligar, S. G.; Nienhaus, G. U. Biophys. J., 1999, 77, 1036-1051.
[11] "CO as a vibrational probe of heme protein active sites", Spiro, T. G.; Wasbotten, I. H. J. Inorg. Biochem., 2005, 99, 34–44.
[12] "Vibrational Analysis of the Model Complex (μ-edt)[Fe(CO)3]2 and Comparison to Iron-Only Hydrogenase: The Activation Scale of Hydrogenase Model Systems", Galinato, M. G. I.; Whaley, C. M.; Lehnert, N. Inorg. Chem., 2010, 49, 3201-3215.. This work was recognized as a cover article in Angewandte Chemie (Figure 3, lower right).
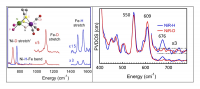
More recently, we have pushed to higher frequencies to observe bands associated with Fe-H or Fe-D motion. In a hydride-bridged model compound, we observed not only Fe-H/D modes but also a characteristic Ni-H-Fe 'wag', and a candidate Ni-H-Fe 'wag' mode was also seen in the reduced Ni-R form of [NiFe] H2ase (Figure 4). This confirms that the hydride in Ni-R is a bridging H, and it eliminates other candidates that involved H bound solely to Ni or Fe.
Figure 3. Top panel: The structures of the Fe sites in [NiFe] H2ase. (a) overall view of the enzyme with 2 subunits, the catalytic Ni-Fe site and the electron transport chain; (b-e) detailed views of individual clusters, including: (b) 'distal' [4Fe-4S] cluster, showing Cys3His ligation, (c) 'medial' [3Fe-4S] cluster, (d) 'proximal' [4Fe-4S], and (e) Ni-Fe active site with amino acid ligands. The atoms are Fe (orange), Ni (green), S (yellow), C (black), N (blue) and O (red)[5][5]"NiFe hydrogenases: structural and spectroscopic studies of the reaction mechanism", Ogata, H.; Lubitz, W.; Higuchi, Y. Dalton Trans., 2009, 37, 7577-7587.. Bottom panel: (left) the NRVS spectrum for NiFe H2ase; (right) the cover story published in Angewandt Chemie International Ed. Volume 52 (2013) –reprint with permit from Angewandt.
Figure 4. Left: Recent data on a model compound showing not only Fe-H/D modes but also Ni-H-Fe bend ('wag'). Right: Observation of candidate Ni-H-Fe 'wag' mode in [NiFe] H2ase.
N2ase Progress
Nitrogenase (N2ase) is the enzyme responsible for biological nitrogen fixation[13-15][13] "Climbing Nitrogenase: Toward a Mechanism of Enzymatic Nitrogen Fixation", Hoffman, B. M.; Dean, D. R.; Seefeldt, L. C. Acc. Chem. Res., 2009, 42, 609-619.
[14] "Mechanism of Mo-Dependent Nitrogenase", Seefeldt, L. C.; Hoffman, B. M.; Dean, D. R. Ann. Rev. Biochem., 2009, 78, 701-722.
[15] "The Evolution and Future of Earth's Nitrogen Cycle", Canfield, D. E.; Glazer, A. N.; Falkowski, P. G. Science, 2010, 330, 192-196.. In the Azotobacter vinelandii (Av) Mo-dependent nitrogenase (N2ase), this catalysis takes place at an active site MoFe7S9 'FeMo cofactor' (Figure 5, upper left). It is now known that N2ase can also produce CxHy hydrocarbons from CO[16-18][16] "Vanadium Nitrogenase Reduces CO", Lee, C. C.; Hu, Y.; Ribbe, M. W. Science, 2010, 329, 642.
[17] "Tracing the Hydrogen Source of Hydrocarbons Formed by Vanadium Nitrogenase", Lee, C. C.; Hu, Y. L.; Ribbe, M. W. Angew. Chem., 2011, 50, 5545-5547.
[18] "Molybdenum Nitrogenase Catalyzes the Reduction and Coupling of CO to Form Hydrocarbons", Yang, Z.-Y.; Dean, D. R.; Seefeldt, L. C. J. Biol. Chem., 2011, 286, 19417–19421. and even CH4 from CO2[19,20][19] "Carbon dioxide reduction to methane and coupling with acetylene to form propylene catalyzed by remodeled nitrogenase", Yang, Z.-Y.; Moure, V. R.; Dean, D. R.; Seefeldt, L. C. Proc. Nat. Acad. Sci., 2012, 109, 19644-19648.
[20] "Nitrogenase reduction of carbon-containing compounds", Seefeldt, L. C.; Yang, Z.-Y.; Duval, S.; Dean, D. R. Biochim Biophys Acta, 2013, 1827, 1102-1111.. We have used NRVS to characterize the effects of CO bonding on the normal modes of the FeMo cofactor.
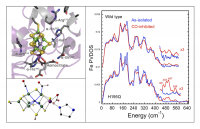
As seen in Figure 5 (right panel), dramatic changes in the NRVS are seen under high-CO conditions, especially in a 188 cm-1 mode associated with symmetric 'breathing' of the [6Fe-X] central cage of the FeMo-cofactor. Similar changes are reproduced with the α-H195Q N2ase variant. In the frequency region above 450 cm-1, additional features are seen that are assigned to Fe-CO bending and stretching modes, and these assignments have been confirmed by observation 13CO isotope shifts. An example of one of our candidate DFT models to explain CO inhibition is illustrated in Figure 5 (lower left).
Figure 5. Top left: structure of the N2ase MoFe protein structure around the FeMo-cofactor for the wild-type enzyme. Bottom left: one of the structures proposed for CO-inhibited wild-type enzyme. Right: 57Fe NRVS for wild-type (top) and H195Q Av (bottom) N2ase under as-isolated (-) and high-CO (-) conditions.
Acknowledgements
This work was funded by NIH GM-65440, NSF CHE-0745353, and the DOE Office of Biological and Environmental Research. These experiments were performed at BL09XU/SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI; Proposal No. 2010B0032-2013A0032).
Publications from this Project
[1] "Characterization of Iron Dinitrosyl Species Formed in the Reaction of Nitric Oxide with a Biological Rieske Center", Tinberg, C. E.; Tonzetich, Z. J.; Wang, H.; Do, L. H.; Yoda, Y.; Cramer, S. P.; Lippard, S. J. J. Am. Chem. Soc., 2010, 132, 18168-18176.
[2] "Fe-H/D stretching and bending modes in nuclear resonant vibrational, Raman and infrared spectroscopies: Comparisons of density functional theory and experiment", Pelmenschikov, V.; Guo, Y.; Wang, H.; Cramer, S. P.; Case, D. A. Faraday Disc., 2011, 148, 409-420.
[3] "Dynamics of the [4Fe-4S] Cluster in Pyrococcus furiosus D14C Ferredoxin via Nuclear Resonance Vibrational and Resonance Raman Spectroscopies, Force Field Simulations, and Density Functional Theory Calculations", Mitra, D.; Pelmenschikov, V.; Guo, Y.; Case, D. A.; Wang, H.; Dong, W.; Tan, M.-L.; Ichiye, T.; Francis E.; Jenney, J.; Adams, M. W. W.; Yoda, Y.; Zhao, J.; Cramer, S. P. Biochemistry, 2011, 50, 5220–5235.
[4] "Characterization of a Synthetic Peroxodiiron(III) Protein Model Complex by Nuclear Resonance Vibrational Spectroscopy", Do, L. H.; Wang, H.; Tinberg, C. E.; Dowty, E.; Yoda, Y.; Cramer, S. P.; Lippard, S. J. Chem. Commun., 2011, 47, 10945-10947.
[5] "EXAFS and NRVS Reveal a Conformational Distortion of the FeMo-cofactor in the MoFe Nitrogenase Propargyl Alcohol Complex", George, S. J.; Barney, B. M.; Mitra, D.; Igarashi, R. Y.; Guo, Y.; Dean, D. R.; Cramer, S. P.; Seefeldt, L. C. J. Inorg. Biochem., 2012, 112, 85-92.
[6] "Nuclear resonance vibrational spectroscopy (NRVS) of rubredoxin and MoFe protein crystals", Guo, Y.; Brecht, E.; Aznavour, K.; Nix, J.; Xiao, Y.; Wang, H.; George, S.; Bau, R.; Keable, S.; Peters, J.; Adams, M. W.; Jr, F. J.; Sturhahn, W.; Alp, E.; Zhao, J.; Yoda, Y.; Cramer, S. P. Hyperfine Interact., 2013, 222, 77-90.
[7] "Real sample temperature: a critical issue in the experiments of nuclear resonant vibrational spectroscopy on biological samples", Wang, H. X.; Yoda, Y.; Kamali, S.; Zhou, Z. H.; Cramer, S. P. J. Synchrotron Rad., 2012, 19, 257-263.
[8] "Observation of the Fe-CN and Fe-CO Vibrations in the Active Site of [NiFe] Hydrogenase by Nuclear Resonance Vibrational Spectroscopy", Kamali, S.; Wang, H.; Mitra, D.; Ogata, H.; Lubitz, W.; Manor, B. C.; Rauchfuss, T. B.; Byrne, D.; Bonnefoy, V.; Jenney Jr., F. E.; Adams, M. W. W.; Yoda, Y.; Alp, E.; Zhao, J.; Cramer, S. P. Angew. Chem. Int. Ed., 2013, 52, 724–728.
[9] "Characterization of [4Fe-4S] Cluster Dynamics and Structure in Nitrogenase Fe Protein at Three Oxidation Levels via Combined NRVS, EXAFS and DFT Analyses", Mitra, D.; George, S. J.; Guo, Y.; Kamali, S.; Keable, S.; Peters, J. W.; Pelmenschikov, V.; Case, D. A.; Cramer, S. P. J. Am. Chem. Soc., 2013, 135, 2530–2543.
[10] "Nuclear Resonance Vibrational Spectroscopy and Electron Paramagnetic Resonance Spectroscopy of 57Fe-Enriched [FeFe] Hydrogenase Indicate Stepwise Assembly of the H-Cluster", Kuchenreuther, J. M.; Guo, Y.; Wang, H.; Myers, W. K.; George, S. J.; Boyke, C. A.; Yoda, Y.; Alp, E. E.; Zhao, J.; Britt, R. D.; Swartz, J. R.; Cramer, S. P. Biochemistry, 2013, 52, 818–826.
[11] "Synchrotron Radiation Based Nuclear Resonant Scattering: Applications to Bioinorganic Chemistry", Guo, Y.; Yoda, Y.; Zhang, X.; Xiao, Y.; Cramer, S. P., in Mössbauer Spectroscopy: Applications in Chemistry, Biology, Industry, and Nanotechnology; Sharma, V. K.; Klingelhofer, G.; Nishida, T.; Ed. Wiley, 2013, 246-271.
References
[1] "Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator", Ding, H. G.; Demple, B. Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 5146-5150.
[2] "Interplay between NO and Fe-S clusters: Relevance to biological systems", Drapier, J. C. Methods, 1997, 11, 319-329.
[3] "X-ray Crystal Structure of the Fe-Only Hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom Resolution", Peters, J. W.; Lanzilotta, W. N.; Lemon, B. J.; Seefeldt, L. C. Science, 1998, 282, 1853-1858.
[4] "Nuclear Resonance Vibrational Spectroscopy and Electron Paramagnetic Resonance Spectroscopy of 57Fe-Enriched [FeFe] Hydrogenase Indicate Stepwise Assembly of the H-Cluster", Kuchenreuther, J. M.; Guo, Y.; Wang, H.; Myers, W. K.; George, S. J.; Boyke, C. A.; Yoda, Y.; Alp, E. E.; Zhao, J.; Britt, R. D.; Swartz, J. R.; Cramer, S. P. Biochemistry, 2013, 52, 818–826.
[5] "NiFe hydrogenases: structural and spectroscopic studies of the reaction mechanism", Ogata, H.; Lubitz, W.; Higuchi, Y. Dalton Trans., 2009, 37, 7577-7587.
[6] "Biological Activation of Hydrogen", Happe, R. P.; Roseboom, W.; Pierik, A. J.; Albracht, S. P. J.; Bagley, K. A. Nature, 1997, 385, 126.
[7] "Quantitative Vibrational Dynamics of Iron in Carbonyl Porphyrins", Leu, B. M.; Silvernail, N. J.; Zgierski, M. Z.; Wyllie, G. R. A.; Ellison, M. K.; Scheidt, W. R.; Zhao, J.; Sturhahn, W.; Alp, E. E.; Sage, J. T. Biophys. J., 2007, 92, 3764-3783.
[8] "Carbonyl Tilting and Bending Potential Energy Surface of Carbon Monoxyhemes", Ghosh, A.; Bocian, D. F. J. Phys. Chem., 1996, 100, 6363-6367.
[9] "Is Bound CO Linear or Bent in Heme-Proteins - Evidence from Resonance Raman and Infrared Spectroscopic Data", Li, X. Y.; Spiro, T. G. J. Am. Chem. Soc., 1988, 110, 6024-6033.
[10] "Connection between the taxonomic substates and protonation of histidines 64 and 97 in carbonmonoxy myoglobin", Müller, J. D.; McMahon, B. H.; Chien, E. Y. T.; Sligar, S. G.; Nienhaus, G. U. Biophys. J., 1999, 77, 1036-1051.
[11] "CO as a vibrational probe of heme protein active sites", Spiro, T. G.; Wasbotten, I. H. J. Inorg. Biochem., 2005, 99, 34–44.
[12] "Vibrational Analysis of the Model Complex (μ-edt)[Fe(CO)3]2 and Comparison to Iron-Only Hydrogenase: The Activation Scale of Hydrogenase Model Systems", Galinato, M. G. I.; Whaley, C. M.; Lehnert, N. Inorg. Chem., 2010, 49, 3201-3215.
[13] "Climbing Nitrogenase: Toward a Mechanism of Enzymatic Nitrogen Fixation", Hoffman, B. M.; Dean, D. R.; Seefeldt, L. C. Acc. Chem. Res., 2009, 42, 609-619.
[14] "Mechanism of Mo-Dependent Nitrogenase", Seefeldt, L. C.; Hoffman, B. M.; Dean, D. R. Ann. Rev. Biochem., 2009, 78, 701-722.
[15] "The Evolution and Future of Earth's Nitrogen Cycle", Canfield, D. E.; Glazer, A. N.; Falkowski, P. G. Science, 2010, 330, 192-196.
[16] "Vanadium Nitrogenase Reduces CO", Lee, C. C.; Hu, Y.; Ribbe, M. W. Science, 2010, 329, 642.
[17] "Tracing the Hydrogen Source of Hydrocarbons Formed by Vanadium Nitrogenase", Lee, C. C.; Hu, Y. L.; Ribbe, M. W. Angew. Chem., 2011, 50, 5545-5547.
[18] "Molybdenum Nitrogenase Catalyzes the Reduction and Coupling of CO to Form Hydrocarbons", Yang, Z.-Y.; Dean, D. R.; Seefeldt, L. C. J. Biol. Chem., 2011, 286, 19417–19421.
[19] "Carbon dioxide reduction to methane and coupling with acetylene to form propylene catalyzed by remodeled nitrogenase", Yang, Z.-Y.; Moure, V. R.; Dean, D. R.; Seefeldt, L. C. Proc. Nat. Acad. Sci., 2012, 109, 19644-19648.
[20] "Nitrogenase reduction of carbon-containing compounds", Seefeldt, L. C.; Yang, Z.-Y.; Duval, S.; Dean, D. R. Biochim Biophys Acta, 2013, 1827, 1102-1111.