Volume 18, No.4 Pages 282 - 286
1. 最近の研究から/FROM LATEST RESEARCH
Elucidation of Non-heme Iron Enzyme Intermediates Using Nuclear Resonance Vibrational Spectroscopy
[1]Department of Chemistry, Stanford University, [2]Research & Utilization Division, JASRI
- Abstract
- Mononuclear and binuclear non-heme iron enzymes are found in numerous aerobic organisms and perform an array of different chemistries, which are significant in the development of drugs and industrial catalysts. The goal of this research is to understand mechanisms by which the iron cofactors of these enzymes activate O2 to perform the diverse chemistries. Recently, the first structural elucidation of the key reactive Fe(IV)=O intermediate of a mononuclear non-heme iron enzyme SyrB2 has been achieved by utilizing nuclear resonance vibrational spectroscopy (NRVS), and it has been correlated to the function of the enzyme in combination with density functional theory (DFT) computations. For the similar achievement in elucidating binuclear non-heme iron enzyme intermediates, we also established bases for the NRVS analyses of peroxo-bridged biferric and mono- and di-oxo-bridged high-valent diiron complexes.
Introduction1[1] a) Solomon, E. I. et al., Chem. Rev. 2000, 100, 235.
b) Solomon, E. I. et al., Acc. Chem. Res. 2013, ASAP.
1. Mononuclear non-heme iron enzymes
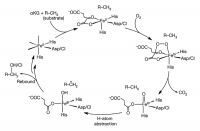
Mononuclear non-heme iron enzymes have an Fe center coordinated by a ligand set called a facial triad, which consists of two histidine and one carboxylate amino acids. This Fe center plays key roles in metabolic pathways, antibiotic synthesis, and bioremediation. Specifically, mononuclearα-ketoglutarate (α-KG)-dependent non-heme iron enzymes utilize the Fe center and another cofactorα-KG to catalyze oxidative reactions for antibiotic synthesis, demethylation of DNA in epigenetics, and the hypoxic response. These enzymes traverse similar steps in their catalytic cycles; their high-spin Fe(II) center activates O2, arriving at the key reactive Fe(IV)=O intermediate (Fig. 1), which can perform hydrogen-atom abstraction for hydroxylation, halogenation, desaturation or electrophillic aromatic substitution (EAS). In halogenase SyrB2 (syringomycin biosynthesis enzyme 2) from Pseudomonas syringae pv. syringae, this Fe(IV)=O intermediate abstracts a hydrogen-atom from the substrate to form an Fe(III)—OH species and a substrate radical. For its native L-threonine (L-Thr) substrate, SyrB2 performs a subsequent Cl•rebound to form 4-Cl-L-Thr; with non-native substrates such as L-norvaline (L-Nva), however, it performs HO•rebound instead, forming a hydroxylated product (Fig. 1, left)2[2] Matthews, M. L. et al., PNAS, 2009, 106, 17723.. To understand this versatile reactivity of the SyrB2 Fe(IV)=O intermediate, its structural elucidation was necessary.
______________________________
* E-mail: edward.solomon@stanford.edu
Figure 1. Catalytic cycle of αKG-dependent non-heme iron enzymes.
αKG and substrate binding induces a six-coordinate to five-coordinate conversion (top), providing a site for O2 to bind and form an Fe(IV)–peroxo species that nucleophilically attacks αKG, producing a peroxo-bridged Fe(IV) species (right). Decarboxylation of αKG leads to the reactive Fe(IV)=O intermediate (bottom right), which goes on to perform hydrogen-atom abstraction (bottom left) and subsequent rebound hydroxylation or halogenation.
2. Binuclear non-heme iron enzymes
Binuclear non-heme iron enzymes have two coupled Fe centers coordinated mostly by two histidine and four carboxylate amino acids, two of which bridge two Fe centers. This binuclear Fe center is observed in numerous enzymes that are essential for aerobic metabolism. For instance, class Ia ribonucleotide reductase (RR) utilizes the binuclear Fe center to generate DNA blocks, and has been a target for anti-cancer drug development as determining the proliferation of human cells. A binuclear Fe center in soluble methane monooxygenase (sMMO) converts methane to methanol and has inspired the development of biofuel catalysts. The high-spin coupled Fe(II)Fe(II) center of these enzymes activate O2 and form peroxo-bridged biferric intermediates (Fig. 2). In RR, the first formed peroxy intermediate P that has characteristic purple color needs to convert to another peroxy intermediate P’, which is colorless3[3] Saleh, L. et al., Biochemistry, 2004, 43, 5953.. Structural differences between P and P’ and why this conversion is necessary for activity are unknown. The P’ intermediate evolves to a highly reactive Fe(III)Fe(IV) intermediate called X in RR, while the purple peroxy intermediate in MMO forms an Fe(IV)2 intermediate Q. These high-valent intermediate X and Q perform thermodynamically challenging H atom abstraction from tyrosine in RR and hydroxylation of methane in MMO. The mechanisms of these reactions remain to be elucidated due to the structural ambiguity of X and Q.
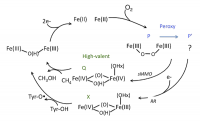
Figure 2. Catalytic cycle of binuclear non-heme iron enzymes.
Upon O2 binding, the biferrous site bridged by two carboxylate amino acids (top) converts to a peroxy-bridged biferric state (right, P/P’) and then a high-valent state (middle, Q for MMO and X for RR). The high-valent state intermediates abstract an H atom from methane or tyrosine and turns to a resting biferric state (left).
Method4[4] Seto, M. et al., Phys. Rev. Lett. 1995, 74, 3828.
Vibrational spectroscopy can provide valuable information on the geometric structures of transient species, but traditional techniques such as resonance Raman (rR) spectroscopy have not been successful in determining the mononuclear Fe(IV)=O intermediate and the binuclear P’, X, and Q intermediates due to their photolability and lack of appropriate absorption bands for resonance enhancement. Therefore, nuclear resonance vibrational spectroscopy (NRVS) is the ideal alternative, because it is not subject to optical rules as probing vibrational side bands of 57Fe nuclear transitions with very bright, tunable X-rays at 14.4 keV. All the vibrational modes that contain significant Fe displacement appear in the NRVS spectra and these modes are sensitive to structural variations and thus allow us to distinguish possible structural candidates. The enrichment of 57Fe in the samples studied and the yield of the intermediate species of interest were ensured by Mössbauer spectroscopy. For NRVS data collection, 1-mm thick samples, either solutions or powders, were placed in a cell made of Lucite and covered with polyimide tape. NRVS data were collected at low temperature below 80 K. Each scan was monitored to ensure the absence of photo-induced damage of samples and multiple scans were accumulated until a reasonable signal-to-noise ratio was obtained. The partial vibrational density-of-states (PVDOS) spectra were obtained by processing accumulated data with PHEONIX program and analyzed using DFT calculations based on known models.
Results and Conclusions
1. Structural Elucidation of SyrB2 Fe(IV)=O intermediate5[5] Wong, S. D. et al., Nature, 2013, 499, 320.
Recently, a sizable amount of an Fe(IV)=O intermediate was trapped in the halogenase SyrB2 and its NRVS structural characterization was performed at the Fe(IV) center coordinated by either Cl– or Br–. Three major NRVS features were observed with the intensity down-shifted in energy upon Cl– to Br– mass perturbation (Fig. 3 top). To elucidate the Fe(IV)=O intermediate structure that can reproduce this NRVS spectral observation, all possible structural candidates were computationally generated by taking SyrB2 from its Fe(II) resting form to the Fe(IV)=O intermediate along the O2 reaction coordinate (Fig. 1). The NRVS spectra of all candidates were predicted using a calibrated DFT models, and among them, only the NRVS spectrum of a five-coordinate, trigonal bipyramidal geometric structure reproduced the experimental NRVS data (Fig. 3) with three features associated with vibrational modes given in Fig. 4.
From reaction coordinate calculations, depending on the type of substrate, this trigonal bipyramidal Fe(IV)=O intermediate is oriented differently. For the native L-Thr substrate, the Fe(IV)=O bond is oriented perpendicular to the substrate C–H bond, while for the non-native L-Nva substrate, it is oriented parallel and pointing towards the substrate C–H bond (Fig. 5 left). For the former case, the H atom of the C–H bond is abstracted by a π-frontier molecular orbital (FMO) of the Fe(IV)=O bond, which results in an Fe(III)—OH intermediate poised for Cl•rebound and halogenation reactivity (Fig. 5 right). Alternatively, for the latter case of the non-native substrate, the Fe(IV)=O intermediate instead utilizes a σ-FMO for H-atom abstraction, forming an Fe(III)—OH intermediate poised for HO•rebound and hydroxylation reactivity (Fig. 5 right). This provides an explanation for the diverse reactivity of SyrB2 depending on the type of substrate.

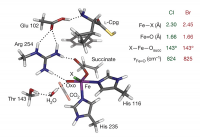
Figure 3. [Top] Experimental NRVS partial vibrational density-of-states spectra of Fe(IV)=O intermediate of SyrB2 ligated by Cl or Br, labeled SyrB2–Cl and SyrB2–Br respectively. Regions 3, 2 and 1 containing intense features are indicated by brackets. [Middle] Density functional theory (DFT)-predicted PVDOS NRVS spectra of (bottom) five-coordinate trigonal bipyramidal structural candidate. All bond lengths in Å.
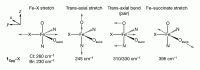
Figure 4. Normal modes of the Fe(IV)=O intermediate.
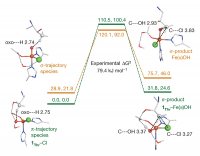
Figure 5. Hydrogen-atom abstraction reaction coordinates.
Energies (given as ΔE/ΔG in kJ mol−1) for the π-trajectory (1Thr–Cl, green numerals and levels) and the σ-trajectory (orange numerals and levels) are shown. Reactants are on the left; energies for the transition state are given at the top; structures of Fe(III)–OH products are displayed on the right, showing the π-product 1Thr–Fe(III)–OH (with hydrogen-bonding interactions indicated) set up for chlorination and the σ-product set up for hydroxylation. Distances are given in Å.
2. NRVS analytic bases for the structural characterization of P’, X, and Q6[6] a) Park, K. et al., Angew. Chem. Int. Ed. 2013, 52, 1294.
b) Park, K. et al., PNAS, 2013, 110, 6275.
In the case of binuclear non-heme iron species, two Fe centers are bridged and thus their vibrations involve coupled motions. Given this complexity, building correlations between the NRVS spectral features and geometric structures of the 2Fe center is necessary to distinguish structural candidates for peroxy- and high-valent intermediates of binuclear non-heme iron enzymes. We established the correlations by utilizing structurally well-characterized model complexes shown in Figs. 6 & 7. Major NRVS features were assigned, and their sensitivity to the oxidation and spin states of Fe centers and the type and geometric variation of bridging ligands was tested computationally. Based on the correlations found, a library of the NRVS spectra for possible intermediate structures was generated, confirming the capability of NRVS to elucidate the structures of P’, X, and Q intermediates and thus their reactivity in our future studies of enzyme intermediates in RR, MMO, and other binuclear non-heme Fe enzymes.
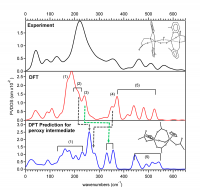
Figure 6. Peroxy-bridged biferric complexes.
The NRVS spectrum of the peroxo, hydroxo biferric complex (top) was assigned with DFT calculation (middle) and extended to predict the NRVS spectrum of the P intermediate. Peroxy out-of-plane and in-plane bending motions (2~4) are sensitive to variations in the Fe-O-O-Fe dihedral angle and the core planarity (i.e. the angle between two planes, each of which is composed of 2Fe and the bridging ligand).

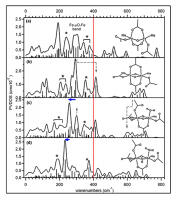
Figure 7. Mono- and di-oxo bridged high-valent diiron complexes.
The DFT models that reproduce the NRVS spectra (top left) were used to predict the NRVS spectra of their high-spin analogs (top right) and then extended to predict the NRVS spectra of possible Q structures (bottom).
Acknowledgments
Funding for this work was provided by the National Institutes of Health (GM-40392) and the National Science Foundation (MCB-0919027). Synchrotron experiments were performed with the approval of the Japan Synchrotron Radiation Research Institute (JASRI; proposal no. 2010B1569 and 2011A1326).
References
[1] a) Solomon, E. I. et al., Chem. Rev. 2000, 100, 235. b) Solomon, E. I. et al., Acc. Chem. Res. 2013, ASAP.
[2] Matthews, M. L. et al., PNAS, 2009, 106, 17723.
[3] Saleh, L. et al., Biochemistry, 2004, 43, 5953.
[4] Seto, M. et al., Phys. Rev. Lett. 1995, 74, 3828.
[5] Wong, S. D. et al., Nature, 2013, 499, 320.
[6] a) Park, K. et al., Angew. Chem. Int. Ed. 2013, 52, 1294. b) Park, K. et al., PNAS, 2013, 110, 6275.