Volume 16, No.3 Pages 197 - 200
1. 最近の研究から/FROM LATEST RESEARCH
Long-term Proposal Report: Structural Study of Regulated Intramembrane Proteolysis
Center for Structural Biology, School of Medicine and School of Life Sciences, Tsinghua University
- Abstract
- Regulated Intramembrane Proteolysis (RIP) is a highly conserved signaling mechanism, where a signaling molecule is cleaved within the lipid bilayer by an intramembrane protease. Traditional wisdom argued that proteolysis requires water; however, in RIP, both the protease and the substrate are integral membrane proteins and the cleavage occurs within the hydrophobic lipid bilayer. It has been most intriguing to scientists how water molecules and substrate get access to the active site of an intramembrane protease. Based on the function and predicted active site, the characterized intramembrane proteases are classified into 4 families: the metalloprotease site-2 protease (S2P), the serine protease Rhomboid, the aspartyl proteases Signal Peptide Peptidase (SPP) and Presenilin. Intramembrane proteases play important roles in a wide range of cellular functions. For example, S2P is a key player in sterol metabolism in cells; Rhomboid works in the Wnt signaling pathway; Presenilin is the most notorious intramembrane protease as it directly cleaves Amyloid Precursor Protein (APP) and results in the accumulation of β-amyloid peptide, the direct pathogen for Alzheimer’s disease. In order to understand the working mechanism of RIP, it requires high-resolution structures of the intramembrane proteases both in the apo-form and in the substrate or inhibitor-bound forms. Successful determination of the proposed structures will also provide invaluable therapeutic potentials to fight deleterious diseases, such as Alzheimer’s disease and cardiovascular diseases. There were a number of important questions that we would like to address. In particular, we would like to understand what regulates the substrate access to the active site of the intramembrane proteases; what are the structure and function mechanism of SPP; and what are the structure and function mechanism of Presenilin? The proposed research aimed to address the above questions.
Introduction
Proteolysis, as the name indicated, refers to the cleavage of protein in the presence of water. However, a new concept of proteolysis was coined in the last two decades, that is, the Regulated Intramembrane Proteolysis (RIP)[1,2][1] Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89 331-340 (1997).
[2] Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100 391-398 (2000).. RIP is a highly conserved and unique signaling mechanism, where both the protease and the substrate are integral membrane proteins and the cleavage occurs within the hydrophobic lipid bilayer[3][3] Jung, K. M. et al. Regulated intramembrane proteolysis of the p75 neurotrophin receptor modulates its association with the TrkA receptor. J. Biol. Chem. 278 42161-42169 (2003).. Based on the function and predicted active site, the characterized intramembrane proteases are classified into 4 families: the metalloprotease site-2 protease (S2P), the serine protease Rhomboid, the aspartyl proteases Signal Peptide Peptidase (SPP) and Presenilin. Intramembrane proteases play important roles in a wide range of cellular functions. For example, S2P is a key player in sterol metabolism in cells; Rhomboid works in the Wnt signaling pathway; Presenilin is the most notorious intramembrane protease as it directly cleaves Amyloid Precursor Protein (APP) and results in the accumulation of β-amyloid peptide, the direct pathogen for Alzheimer’s disease.
In order to understand the working mechanism of RIP, it requires high-resolution structures of the intramembrane proteases both in the apo-form and in the substrate or inhibitor-bound forms. Successful determination of the proposed structures will also provide invaluable therapeutic potentials to fight deleterious diseases, such as Alzheimer’s disease and cardiovascular diseases. For this reason, intense efforts have been invested to the structural determination of intramembrane proteases. The breakthrough was finally made in 2006. Four independent groups successfully solved the structures of the bacterial homologs of Rhomboid in their apo forms[4-7][4] Wang, Y., Zhang, Y. & Ha, Y. Crystal structure of a rhomboid family intramembrane protease. Nature 444 179-180 (2006).
[5] Wu, Z. et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nature Struct. Mol. Biol. 13 1084-1091 (2006).
[6] Lemieux, M. J., Fischer, S. J., Cherney, M. M., Bateman, K. S. & James, M. N. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. USA 104 750-754 (2007).
[7] Ben-Shem, A., Fass, D. & Bibi, E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. USA 104 462-466 (2007).. At the end of year 2007, the structure of an archaebacterial S2P homolog was determined by Dr. Yigong Shi’s group at Princeton University[8][8] Feng, L. et al. Structure of a site-2 protease family intramembrane metalloprotease. Science 318 1608-1612 (2007).. The structures answered the question of how water molecules get access to the active site and provided clue to understanding substrate entry; however, the following questions remain unknown:
1. What regulates the substrate access to the active site?
2. What are the structure and functional mechanism of SPP?
3. What are the structure and functional mechanism of Presenilin?
To address these important questions, we proposed to:
1. To understand the regulatory mechanism of S2P
2. To determine the structure of SPP;
3. To determine the high-resolution structure of Presenilin;
We would like to combine structural biology, biochemistry and other biophysical approaches to understand the working mechanism of RIP.
Despite of the ambitious plan, we understand very well the intrinsic difficulty to deal with membrane proteins. Thus, for a long-term proposal with SPring-8, we anticipated to make major progress on the mechanistic understanding of S2P, and to make preliminary result on the structural study of SPP and presenilin.
Progress
S2P, as the named indicated “site 2 protease”, cleaves substrate only after S1P (site 1 protease cleavage). Why is Site 1 cleavage required for S2P function? This remains to be one of the central questions in the understanding of the regulation of intramembrane proteolysis.
Dr. Yigong Shi’s group at the Center for Structural Biology, Tsinghua University, combined biochemistry and structural biology to address this question by focusing on the bacterial ortholog of S2P, RseP (also known as YaeL). In response to accumulation of unfolded outer membrane proteins (OMP) in the envelope of Escherichia coli, RseP (also known as YaeL), a representative member of the S2P proteases, cleaves a membrane-anchored protein RseA at an intramembrane site that is proximal to the cytoplasm (Fig. 1). Because the amino-terminal sequence of RseA is bound to the transcription factor sE, this cleavage results in the release of the amino-terminal RseA-sE complex into the cytoplasm, where RseA is selectively degraded by proteases and the freed sE factor activates transcription of genes that cope with envelope stress. As is the case for all characterized S2Ps, RseP can cleave RseA only after a prior cleavage mediated by the membrane-anchored protease DegS, a known S1P[9][9] Alba, B. M., Leeds, J. A., Onufryk, C., Lu, C. Z. & Gross, C. A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)- dependent extracytoplasmic stress response. Genes Dev. 16 2156-2168 (2002).. DegS is a serine protease with a carboxy-terminal PDZ domain and an amino-terminal transmembrane segment (Fig. 1). In the absence of envelope stress, DegS exists in an auto-inhibited state. Binding of the C-terminal residues of unfolded OMPs to the PDZ domain of DegS triggers its allosteric activation[10,11][10] Walsh, N. P., Alba, B. M., Bose, B., Gross, C. A. & Sauer, R. T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113 61-71 (2003).
[11] Sohn, J., Grant, R. A. & Sauer, R. T. Allosteric activation of DegS, a stress sensor PDZ protease. Cell 131 572-583 (2007)., which subsequently cleaves the periplasmic domain of RseA between amino acids Val148 and Ser149 (Fig. 1)[12][12] Cezairliyan, B. O. & Sauer, R. T. Inhibition of regulated proteolysis by RseB. Proc. Natl. Acad. Sci. USA 104 3771-3776 (2007). .

Figure 1 A schematic diagram of the proteolytic cascade across the inner membrane of Gram-negative bacteria.
In the study, they reported three novel findings. First, they demonstrated that identity of the newly exposed carboxy-terminal residue of RseA, as a result of Site-1 cleavage by DegS, determines whether Site-2 cleavage can occur and the extent of Site-2 cleavage. Conserved mutation of Val148 (to Ala, Ile, Leu, Cys, Thr, and Asn) allowed both Site-1 and Site-2 cleavages. Second, they showed that mutation of residues in the putative peptidebinding groove of RseP PDZ domains abolished its activity to mediate the Site-2 cleavage. These two observations strongly implicate a mechanism whereby direct interaction of the newly exposed carboxy-terminus of RseA with the PDZ domains of RseP leads to the activation of RseP and subsequent Site-2 cleavage. Interestingly, however, such interactions eluded detection. This apparent dilemma was explained by the third finding – the putative peptide-binding grove of the first PDZ domain is blocked by a structural element that appear to be conserved among the PDZ domains of S2P.
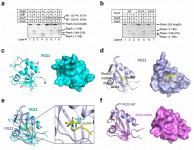
The results were published as a research article in PNAS[13][13] Li, X. et al. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. USA 106 14837-14842 (2009). .
Future plan:
We are now continuing our effort on the structural determination of SPP and presenilin. We have successfully set up the protein expression and purification system and obtained reasonable yield of the recombinant proteins. We anticipate some breakthrough on these two important proteins in the near future.
Annotation:
The figures and some of the progress description were adopted from the published paper by Li et al[13][13] Li, X. et al. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. USA 106 14837-14842 (2009)..
References:
[1] Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89 331-340 (1997).
[2] Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100 391-398 (2000).
[3] Jung, K. M. et al. Regulated intramembrane proteolysis of the p75 neurotrophin receptor modulates its association with the TrkA receptor. J. Biol. Chem. 278 42161-42169 (2003).
[4] Wang, Y., Zhang, Y. & Ha, Y. Crystal structure of a rhomboid family intramembrane protease. Nature 444 179-180 (2006).
[5] Wu, Z. et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nature Struct. Mol. Biol. 13 1084-1091 (2006).
[6] Lemieux, M. J., Fischer, S. J., Cherney, M. M., Bateman, K. S. & James, M. N. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. USA 104 750-754 (2007).
[7] Ben-Shem, A., Fass, D. & Bibi, E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. USA 104 462-466 (2007).
[8] Feng, L. et al. Structure of a site-2 protease family intramembrane metalloprotease. Science 318 1608-1612 (2007).
[9] Alba, B. M., Leeds, J. A., Onufryk, C., Lu, C. Z. & Gross, C. A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)- dependent extracytoplasmic stress response. Genes Dev. 16 2156-2168 (2002).
[10] Walsh, N. P., Alba, B. M., Bose, B., Gross, C. A. & Sauer, R. T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113 61-71 (2003).
[11] Sohn, J., Grant, R. A. & Sauer, R. T. Allosteric activation of DegS, a stress sensor PDZ protease. Cell 131 572-583 (2007).
[12] Cezairliyan, B. O. & Sauer, R. T. Inhibition of regulated proteolysis by RseB. Proc. Natl. Acad. Sci. USA 104 3771-3776 (2007).
[13] Li, X. et al. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. USA 106 14837-14842 (2009).








