Volume7 No.2
SPring-8 Section A: Scientific Research Report
Structural Studies of Orotate Phosphoribosyl Transferase (TTHA1742) from Hyperthermophilic Thermus thermophilus HB8
Alagappa University
- Abstract
-
The enzyme orotate phosphoribosyl transferase (OPRTase) is involved in the de novo pyrimidine nucleotide biosynthesis; it is a good target for antimicrobial agents. Also, some of the cancerous cells are known to depend on de novo pathway, this enzyme could be a potential drug target for cancer therapy. Orotate phosphoribosyl transferase is an important enzyme, which converts orotate to orotate monophosphate in the fifth step of pyrimidine biosynthesis. As the enzyme is from a hyperthermophile it is known to be stable at high temperatures and can be used as a model organism. Based on the importance of the enzyme, OPRTase from the extreme thermophile, Thermus thermophilus HB8 has been cloned, purified by ion exchange and size exclusion chromatography (SEC). Very tiny crystals appeared in the condition 0.2 M calcium acetate hydrate, 0.1 M sodium cacodylate trihydrate pH 6.5 and 18% w/v polyethylene glycol 8,000.
キーワード: Orotate phosphoribosyl transferase; cancer therapy; de novo pathway
Introduction
Six enzymatic reactions are involved in the conversion of glutamine into uridine monophosphate (UMP) of pyrimidine biosynthesis. In the fifth committed step of de novo pyrimidine biosynthesis, OPRTase catalyzes the transfer of ribosyl phosphate group from 5-phosphoribose 1-diphosphate to Orotate molecule that leads to the formation of Orotidine Mono Phosphate (OMP) [1,2]. OPRTase belongs to the family of phosphoribosyltransferases (PRTases) involved in both de novo and salvage pathways of nucleotide synthesis as well as in the biosynthesis of arginine and proline [3,4]. OPRTase is found to be associated in nucleotide metabolism and clinically important because of an inborn defect of the enzyme linked to hereditary orotic aciduria that may result in mental retardation [5-8].
Structural insights of OPRTase will provide the prolific information in designing a drug molecule against pathogenesis and in similar way it is applicable to frame the strategies in targeting the pathogenic microorganisms pyrimidine pathway.
To determine the crystal structure of the protein and subsequently carry out complex crystals experiments by exploring the chemical space as an effort to find the suitable inhibitor for the enzymatic activity. To perform some protein stability and enzyme activity studies at different temperatures.
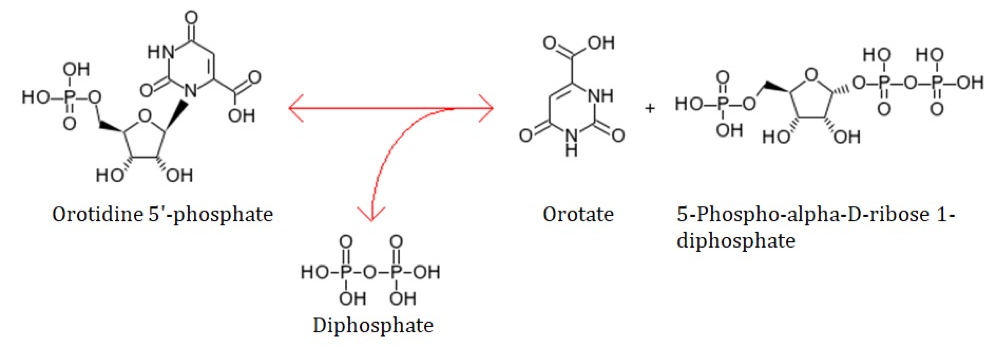
Figure 1. Enzymatic reaction of OPRTase.
Materials and methods
Expression and Purification
E. coli BL21 cells were transformed with the recombinant pET-11a plasmid and grown at 37° C in LB medium containing ampicillin (100 mg ml-1) to an OD of 0.6 at 600 nm. Subsequently, protein expression was induced by adding 0.3 mM isopropyl-d-1-thiogalactopyranoside (IPTG). The cells were grown for an additional 3-4 h. The bacterial cells were collected by centrifugation and disrupted by sonication in 50 mM Tris-HCl buffer pH 7.5 that contained 20 mM NaCl, 5% glycerol with the presence of 1 mM PMSF, 1 mM Benzamidine and 1 mM DTT. The soluble fraction from centrifugation at 15000 rpm for 1 h was heated at 80˚ C for 15 min. The denatured proteins were removed by centrifugation at 15000 rpm for 1 h. The protein supernatant was loaded onto a column Q Sepharose FF (GE Healthcare, USA) that was pre-equilibrated in 50 mM Tris pH-7.5, 20 mM NaCl and 5% glycerol. The protein fractions were eluted using a linear gradient from 50 mM to 500 mM NaCl. The purity of the fractions was analyzed by SDS-PAGE and fractions containing the protein were pooled and concentrated. The concentrated protein molecular weight was checked by using MALDI-TOF.
Crystallization
The highly purified TtOPRTase enzyme was concentrated to 20 mg ml-1 using centrifugal filters Ultracel-30K (Merck Millipore, USA). Preliminary crystallization screening was performed using the microbatch method of crystallization at 293 K. Initial crystallization screening was carried using the Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2 (Hampton Research, USA) screens. All drops were equilibrated with 1 μl of TtOPRTase at a concentration of approximately 20 mg/ml and 1 μl crystallization solution with 10 μl Al’s oil. Later, manual optimization of the crystal-growth conditions was performed using the hanging-drop vapor-diffusion method in 24-well VDX plates (Hampton Research, USA).
Cryoprotection and X-ray Diffraction
Crystals soaked in cryoprotectant solution [0.2 M calcium acetate hydrate, 0.1 M sodium cacodylate trihydrate pH 6.5 and 18% w/v polyethylene glycol 8,000 with 20% (v/v) glycerol] for 30 s and mounted on a nylon loop, flash-cooled at 100 K using a nitrogen stream and the crystals were checked for diffraction using X-ray wavelength of 0.9 Å on beamline BL44XU at SPring-8, Japan.
Results and Discussion
Crystallization
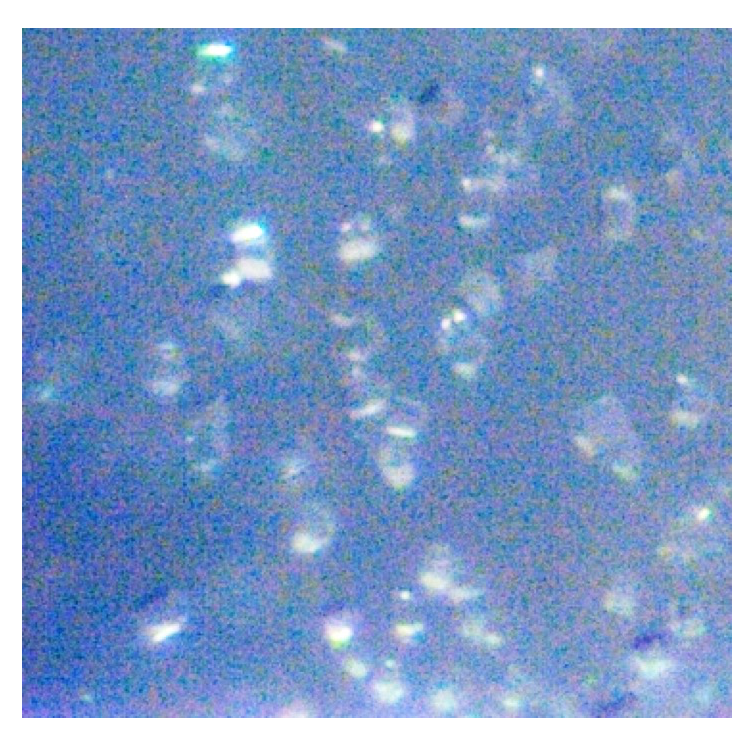
Initial crystallization experiments were carried out at 293 K for TtOPRTase protein by hanging-drop vapor-diffusion and microbatch methods using Crystal Screen 1 and 2, and PEG/Ion Screen (Hampton Research, USA). All drops were equilibrated with 1 μl of TtOPRTase at a concentration of approximately 20 mg ml-1 and 1 μl crystallization solution with 10 μl Al’s oil. Tiny crystals appeared in different conditions of which 0.2 M Calcium acetate hydrate, 0.1 M Sodium cacodylate trihydrate pH 6.5 and 18% w/v Polyethylene glycol 8,000 resulted in microcrystal (Fig. 2). Manual optimization of the crystal-growth conditions were performed using the hanging-drop vapor-diffusion method in 24-well VDX plates (Hampton Research, USA). In further trials, protein concentration was increased to 30 mg ml-1 and crystallization screening using various crystallization methods to produce good quality and diffractable crystals are under progress.
Figure 2. Crystals appeared for TtOPRTase with 0.2 M calcium acetate hydrate, 0.1 M sodium cacodylate trihydrate pH 6.5 and 18% w/v Polyethylene glycol 8,000. The size of the crystals were measured to be 10 µm.
Data Collection at Beamline BL44XU at SPring-8
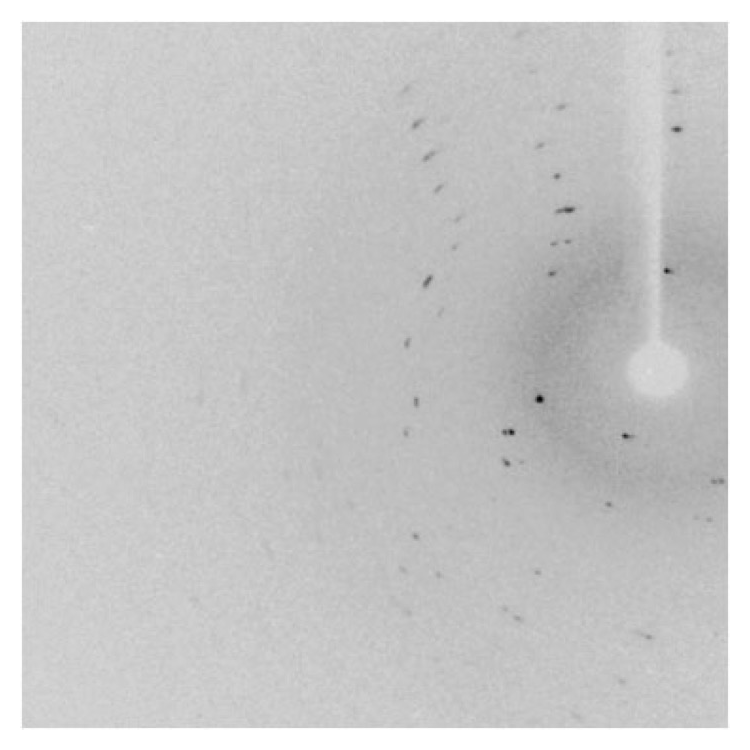
Crystals were soaked in cryoprotectant solution [0.2 M Calcium acetate hydrate, 0.1 M sodium cacodylate trihydrate pH 6.5 and 18% w/v polyethylene glycol 8,000 with 20% (v/v) glycerol] for 30 s, mounted on a nylon loop and flash-cooled at 100 K using a nitrogen stream. Data were collected using X-ray wavelength at 0.9 Å in the beamline BL44XU, SPring-8, Japan for the orotate phosphoribosyl transferase (TTHA1742) protein from hyperthermophilic Thermus thermophilus HB8. The quality of the crystal diffraction obtained was very poor which is shown is Fig. 3 and there is no diffraction pattern in the second image. We observed that the crystals got damaged by radiation immediately. Further, we checked many crystals for diffraction but no diffraction occurred. Hence, we are not able to proceed further measurements. We have selected a new vector (pET-30a+) and the purification strategies are modified, which will lead us to obtain the high diffraction quality crystals in the near future.
Figure 3. Diffraction pattern obtained for TtOPRTase.
Reference
[1] P. F. Coleman, D. P. Suttle, and G. R. Stark, J. Biol. Chem. 252, 6379-6385 (1977).
[2] S. Condon, J. K. Collins, and G. A. O'Donovan, J. Gen. Microbiol. 92, 375-383 (1976).
[3] H. D. Doremus, and A. T. Jagendorf, Plant Physiol. 79, 856–861 (1985).
[4] S. E. Hager, and M. E. Jones, J. Biol. Chem. 242, 5674- 5680 (1967).
[5] H. -H. Hoffmann, et al., Proc. Natl. Acad. Sci. U. S. A. 108, 5777–5782 (2011).
[6] J. Y. Hutson, and M. Downing, J. Bacteriol. 96, 1249-1254 (1968).
[7] M. E. Jones, Annu. Rev. Biochem. 49, 253-279 (1980).
[8] S. R. Krungkrai et al., Biochemistry 44, 1643–1652 (2005).
(Received: August 10, 2018; Accepted: July 16, 2019; Published: August 29, 2019)